BLASTX 2.2.6 [Apr-09-2003]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
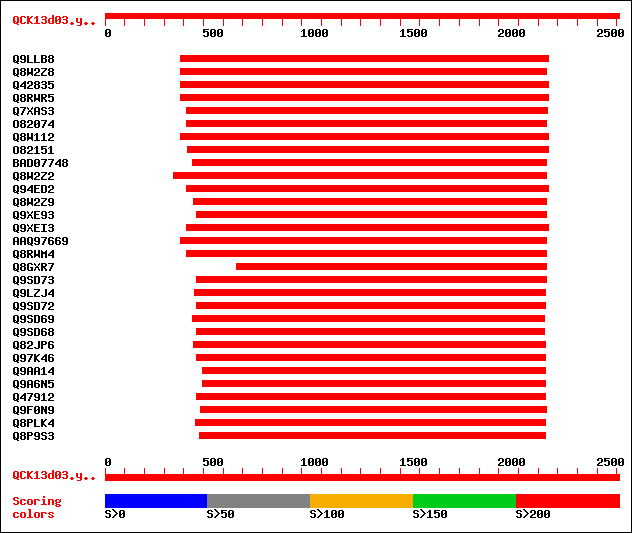
Query= QCK13d03.yg.3.1
(2611 letters)
Database: swall
1,381,838 sequences; 439,479,560 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
sptr|Q9LLB8|Q9LLB8 Exoglucanase precursor. 1231 0.0
sptr|Q8W2Z8|Q8W2Z8 Putative exoglucanase. 1090 0.0
sptr|Q42835|Q42835 Beta-D-glucan exohydrolase, isoenzyme ExoII (... 1077 0.0
sptr|Q8RWR5|Q8RWR5 Beta-D-glucan exohydrolase. 1077 0.0
sptr|Q7XAS3|Q7XAS3 Beta-D-glucosidase. 955 0.0
sptr|O82074|O82074 Beta-D-glucosidase precursor (EC 3.2.1.21). 928 0.0
sptr|Q8W112|Q8W112 Beta-D-glucan exohydrolase-like protein. 924 0.0
sptr|O82151|O82151 Beta-D-glucan exohydrolase. 914 0.0
sptrnew|BAD07748|BAD07748 Putative beta-D-glucan exohydrolase. 907 0.0
sptr|Q8W2Z2|Q8W2Z2 Putative exohydrolase. 903 0.0
sptr|Q94ED2|Q94ED2 Putative beta-glucosidase. 895 0.0
sptr|Q8W2Z9|Q8W2Z9 Unnamed protein product. 890 0.0
sptr|Q9XE93|Q9XE93 Exhydrolase II. 887 0.0
sptr|Q9XEI3|Q9XEI3 Beta-D-glucan exohydrolase isoenzyme ExoI. 873 0.0
sptrnew|AAQ97669|AAQ97669 Beta-glucanase. 870 0.0
sptr|Q8RWM4|Q8RWM4 Beta-glucosidase-like protein. 850 0.0
sptr|Q8GXR7|Q8GXR7 Hypothetical protein. 764 0.0
sptr|Q9SD73|Q9SD73 Beta-D-glucan exohydrolase-like protein (At3g... 716 0.0
sptr|Q9LZJ4|Q9LZJ4 Beta-D-glucan exohydrolase-like protein. 703 0.0
sptr|Q9SD72|Q9SD72 Beta-D-glucan exohydrolase-like protein. 697 0.0
sptr|Q9SD69|Q9SD69 Beta-D-glucan exohydrolase-like protein. 688 0.0
sptr|Q9SD68|Q9SD68 Beta-D-glucan exohydrolase-like protein. 677 0.0
sptr|Q82JP6|Q82JP6 Putative glycosyl hydrolase. 551 e-155
sptr|Q97K46|Q97K46 Beta-glucosidase family protein. 473 e-132
sptr|Q9AA14|Q9AA14 1,4-beta-D-glucan glucohydrolase D. 452 e-125
sptr|Q9A6N5|Q9A6N5 1,4-beta-D-glucan glucohydrolase D. 441 e-122
sptr|Q47912|Q47912 1,4-B-D-glucan glucohydrolase. 438 e-121
sptr|Q9F0N9|Q9F0N9 Cellobiase CelA precursor. 426 e-118
sptr|Q8PLK4|Q8PLK4 Glucan 1,4-beta-glucosidase. 409 e-113
sptr|Q8P9S3|Q8P9S3 Glucan 1,4-beta-glucosidase. 409 e-112
>sptr|Q9LLB8|Q9LLB8 Exoglucanase precursor.
Length = 622
Score = 1231 bits (3184), Expect = 0.0
Identities = 609/622 (97%), Positives = 609/622 (97%)
Frame = +2
Query: 386 MASVHKATTLVLMFCLLALGRAEYLKYKDPKQPVAVRIKDLLGRMTLAEKIGQMTQIERE 565
MASVHKATTLVLMFCLLALGRAEYLKYKDPKQPVAVRIKDLLGRMTLAEKIGQMTQIERE
Sbjct: 1 MASVHKATTLVLMFCLLALGRAEYLKYKDPKQPVAVRIKDLLGRMTLAEKIGQMTQIERE 60
Query: 566 NATADALAKYFIGSVLSGGGSVXXXXXXXXXXXXMVTEMQKGALSTRLGIPIIYGIDAVH 745
NATADALAKYFIGSVLSGGGSV MVTEMQKGALSTRLGIPIIYGIDAVH
Sbjct: 61 NATADALAKYFIGSVLSGGGSVPAPQASAQAWAAMVTEMQKGALSTRLGIPIIYGIDAVH 120
Query: 746 GHNNVYKATIFPHNVGLGATRDPDLVKRIGEATALEVRATGIPYAFAPCIAVCRDPRWGR 925
GHNNVYKATIFPHNVGLGATRDPDLVKRIGEATALEVRATGIPYAFAPCIAVCRDPRWGR
Sbjct: 121 GHNNVYKATIFPHNVGLGATRDPDLVKRIGEATALEVRATGIPYAFAPCIAVCRDPRWGR 180
Query: 926 CYESYSEDPKVVQSLTSLISGLQGDAPADSAGRPYVGGSKKVAACAKHYVGDGGTHNGIN 1105
CYESYSEDPKVVQSLTSLISGLQGDAPADSAGRPYVGGSKKVAACAKHYVGDGGTHNGIN
Sbjct: 181 CYESYSEDPKVVQSLTSLISGLQGDAPADSAGRPYVGGSKKVAACAKHYVGDGGTHNGIN 240
Query: 1106 ENNTIIDTHGLLSIHMPPYYNSIIRGVSTVMVSYSSWNGVKMHANHFLVTDFLKNKLKFR 1285
ENNTIIDTHGLLSIHMPPYYNSIIRGVSTVMVSYSSWNGVKMHANHFLVTDFLKNKLKFR
Sbjct: 241 ENNTIIDTHGLLSIHMPPYYNSIIRGVSTVMVSYSSWNGVKMHANHFLVTDFLKNKLKFR 300
Query: 1286 GFVISDWEGIDRITTPPHANYSYSIEAGVGAGIDMIMVPFRYTEFIDDLTTQVQNKVIPM 1465
GFVISDWEGIDRITTPPHANYSYSIEAGVGAGIDMIMVPFRYTEFIDDLTTQVQNKVIPM
Sbjct: 301 GFVISDWEGIDRITTPPHANYSYSIEAGVGAGIDMIMVPFRYTEFIDDLTTQVQNKVIPM 360
Query: 1466 SRIDDAVYRILRVKFTMGLFENPYPDSSLAGELGKQEHRELAREAVRKSLVLLKNGKSSY 1645
SRIDDAVYRILRVKFTMGLFENPYPDSSLAGELGKQEHRELAREAVRKSLVLLKNGKSSY
Sbjct: 361 SRIDDAVYRILRVKFTMGLFENPYPDSSLAGELGKQEHRELAREAVRKSLVLLKNGKSSY 420
Query: 1646 APLLPLPKKAGKILVAGSHANDLGNQCGGWTITWQGSSGNTTAGTTILSGIEATVDPSTQ 1825
APLLPLPKKAGKILVAGSHANDLGNQCGGWTITWQGSSGNTTAGTTILSGIEATVDPSTQ
Sbjct: 421 APLLPLPKKAGKILVAGSHANDLGNQCGGWTITWQGSSGNTTAGTTILSGIEATVDPSTQ 480
Query: 1826 VVYSESPDSGVLADKYDYAIVVVGEPPYAETFGDNLNLTIPAPGPSVIQSVCGAAKCVVV 2005
VVYSESPDSGVLADKYDYAIVVVGEPPYAETFGDNLNLTIPAPGPSVIQSVCGAAKCVVV
Sbjct: 481 VVYSESPDSGVLADKYDYAIVVVGEPPYAETFGDNLNLTIPAPGPSVIQSVCGAAKCVVV 540
Query: 2006 LISGRPLVVEPYLGDMDALVAAWLPGSEGQGVADVLFGDYGFTGKLPRTWFKSVDQLPMN 2185
LISGRPLVVEPYLGDMDALVA WLPGSEGQGVADVLFGDYGFTGKLPRTWFKSVDQLPMN
Sbjct: 541 LISGRPLVVEPYLGDMDALVATWLPGSEGQGVADVLFGDYGFTGKLPRTWFKSVDQLPMN 600
Query: 2186 VGDAHYDPLFPFGFGLTTKGTK 2251
VGDAHYDPLFPFGFGLTTKGTK
Sbjct: 601 VGDAHYDPLFPFGFGLTTKGTK 622
>sptr|Q8W2Z8|Q8W2Z8 Putative exoglucanase.
Length = 625
Score = 1090 bits (2818), Expect = 0.0
Identities = 530/621 (85%), Positives = 569/621 (91%), Gaps = 2/621 (0%)
Frame = +2
Query: 386 MASVHKATTLVLMFCLLALGRAEYLKYKDPKQPVAVRIKDLLGRMTLAEKIGQMTQIERE 565
M S+HK + ++LM C LG A+Y+KYKDPKQPV+VR+KDLLGRMTLAEKIGQMTQIERE
Sbjct: 1 MGSLHKFSVILLMLCFATLGSAQYVKYKDPKQPVSVRVKDLLGRMTLAEKIGQMTQIERE 60
Query: 566 NATADALAKYFIGSVLSGGGSVXXXXXXXXXXXXMVTEMQKGALSTRLGIPIIYGIDAVH 745
NATA+ +AKYFIGSVLSGGGSV MV EMQKGALSTRLGIP+IYGIDAVH
Sbjct: 61 NATAEQIAKYFIGSVLSGGGSVPAPQASAQAWASMVNEMQKGALSTRLGIPMIYGIDAVH 120
Query: 746 GHNNVYKATIFPHNVGLGATRDPDLVKRIGEATALEVRATGIPYAFAPCIAVCRDPRWGR 925
GHNNVYKATIFPHNVGLGATRDPDLVKRIGEATALEVRATGIPY FAPC+AVCRDPRWGR
Sbjct: 121 GHNNVYKATIFPHNVGLGATRDPDLVKRIGEATALEVRATGIPYVFAPCVAVCRDPRWGR 180
Query: 926 CYESYSEDPKVVQSLTSLISGLQGDAPADSAGRPYVGGSKKVAACAKHYVGDGGTHNGIN 1105
CYESYSEDPKVVQSLT+LISGLQGD P++ GRPYVGGSKKVAACAKHYVGDGGT GIN
Sbjct: 181 CYESYSEDPKVVQSLTTLISGLQGDVPSNDVGRPYVGGSKKVAACAKHYVGDGGTVKGIN 240
Query: 1106 ENNTIIDTHGLLSIHMPPYYNSIIRGVSTVMVSYSSWNGVKMHANHFLVTDFLKNKLKFR 1285
ENNTIIDTHGLL+IHMPPYYNSIIRGVSTVMVSYSSWNGVKMHANH L+TDFLKNKL+FR
Sbjct: 241 ENNTIIDTHGLLTIHMPPYYNSIIRGVSTVMVSYSSWNGVKMHANHHLITDFLKNKLRFR 300
Query: 1286 GFVISDWEGIDRITTPPHANYSYSIEAGVGAGIDMIMVPFRYTEFIDDLTTQVQNKVIPM 1465
GFVISDW+GIDRIT+PPH NYSYSIEAG+GAGIDMIMVP+ YTEFIDDLT QV NK+IPM
Sbjct: 301 GFVISDWQGIDRITSPPHKNYSYSIEAGIGAGIDMIMVPYTYTEFIDDLTEQVNNKIIPM 360
Query: 1466 SRIDDAVYRILRVKFTMGLFENPYPDSSLAGELGKQEHRELAREAVRKSLVLLKNGKSSY 1645
SRIDDAVYRILRVKFTMGLFE+P+ DSSLA ELGKQEHRELAREAVRKSLVLLKNGKSSY
Sbjct: 361 SRIDDAVYRILRVKFTMGLFESPFADSSLADELGKQEHRELAREAVRKSLVLLKNGKSSY 420
Query: 1646 APLLPLPKKAGKILVAGSHANDLGNQCGGWTITWQGSSGNT-TAGTTILSGIEATVDPST 1822
+P+LPLPKKAGKILVAGSHA+DLG QCGGWTITWQG GN TAGTTILS I+ATVDPST
Sbjct: 421 SPVLPLPKKAGKILVAGSHADDLGRQCGGWTITWQGQPGNNITAGTTILSAIKATVDPST 480
Query: 1823 QVVYSESPDSGVL-ADKYDYAIVVVGEPPYAETFGDNLNLTIPAPGPSVIQSVCGAAKCV 1999
VVYSE+PDS V+ DKYDYAIVVVGEPPYAE FGDNLNLTIP PGP+VIQ+VC + KCV
Sbjct: 481 TVVYSENPDSSVVTGDKYDYAIVVVGEPPYAEGFGDNLNLTIPEPGPTVIQTVCKSIKCV 540
Query: 2000 VVLISGRPLVVEPYLGDMDALVAAWLPGSEGQGVADVLFGDYGFTGKLPRTWFKSVDQLP 2179
VVLISGRPLVVEPY+G +DA VAAWLPG+EGQGVADVLFGDYGFTGKL RTWFKSVDQLP
Sbjct: 541 VVLISGRPLVVEPYIGGIDAFVAAWLPGTEGQGVADVLFGDYGFTGKLSRTWFKSVDQLP 600
Query: 2180 MNVGDAHYDPLFPFGFGLTTK 2242
MNVGDAHYDPLFPFG+GLTT+
Sbjct: 601 MNVGDAHYDPLFPFGYGLTTQ 621
>sptr|Q42835|Q42835 Beta-D-glucan exohydrolase, isoenzyme ExoII (EC
3.2.1.58).
Length = 624
Score = 1077 bits (2786), Expect = 0.0
Identities = 528/624 (84%), Positives = 567/624 (90%), Gaps = 2/624 (0%)
Frame = +2
Query: 386 MASVHKATTLVLMFCLLALGRAEYLKYKDPKQPVAVRIKDLLGRMTLAEKIGQMTQIERE 565
M ++HK T ++LMFCL ALG A+YLKYKDPKQP+ VRIKDLLGRMTLAEKIGQMTQIERE
Sbjct: 1 MGNLHKTTFVLLMFCLAALGSADYLKYKDPKQPLGVRIKDLLGRMTLAEKIGQMTQIERE 60
Query: 566 NATADALAKYFIGSVLSGGGSVXXXXXXXXXXXXMVTEMQKGALSTRLGIPIIYGIDAVH 745
NATA+A++KYFIGSVLSGGGSV MV EMQKGALSTRLGIP+IYGIDAVH
Sbjct: 61 NATAEAMSKYFIGSVLSGGGSVPSPQASAAAWQSMVNEMQKGALSTRLGIPMIYGIDAVH 120
Query: 746 GHNNVYKATIFPHNVGLGATRDPDLVKRIGEATALEVRATGIPYAFAPCIAVCRDPRWGR 925
GHNNVYKATIFPHNVGLGATRDP LVKRIGEATALEVRATGIPYAFAPCIAVCRDPRWGR
Sbjct: 121 GHNNVYKATIFPHNVGLGATRDPMLVKRIGEATALEVRATGIPYAFAPCIAVCRDPRWGR 180
Query: 926 CYESYSEDPKVVQSLTSLISGLQGDAPADSAGRPYVGGSKKVAACAKHYVGDGGTHNGIN 1105
CYESYSEDPKVVQS+T+LISGLQGD PA S GRPYVGGSKKVAACAKHYVGDGGT GIN
Sbjct: 181 CYESYSEDPKVVQSMTTLISGLQGDVPAGSEGRPYVGGSKKVAACAKHYVGDGGTFMGIN 240
Query: 1106 ENNTIIDTHGLLSIHMPPYYNSIIRGVSTVMVSYSSWNGVKMHANHFLVTDFLKNKLKFR 1285
EN+TIID HGL++IHMP YYNSIIRGVSTVM SYSSWNG KMHANHFLVTDFLKNKLKFR
Sbjct: 241 ENDTIIDAHGLMTIHMPAYYNSIIRGVSTVMTSYSSWNGKKMHANHFLVTDFLKNKLKFR 300
Query: 1286 GFVISDWEGIDRITTPPHANYSYSIEAGVGAGIDMIMVPFRYTEFIDDLTTQVQNKVIPM 1465
GFVISDW+GIDRIT+PP NYSYS+EAGVGAGIDMIMVPF YTEFIDDLT QV+N +IPM
Sbjct: 301 GFVISDWQGIDRITSPPGVNYSYSVEAGVGAGIDMIMVPFAYTEFIDDLTYQVKNNIIPM 360
Query: 1466 SRIDDAVYRILRVKFTMGLFENPYPDSSLAGELGKQEHRELAREAVRKSLVLLKNGKSSY 1645
SRI+DAVYRILRVKFTMGLFE+PY D SL GELGKQEHR+LAREAVRKSLVLLKNGKS+
Sbjct: 361 SRINDAVYRILRVKFTMGLFESPYADPSLVGELGKQEHRDLAREAVRKSLVLLKNGKSAS 420
Query: 1646 APLLPLPKKAGKILVAGSHANDLGNQCGGWTITWQGSSGN-TTAGTTILSGIEATVDPST 1822
PLLPLPKKAGKILVAGSHA+DLGNQCGGWTITWQG +GN TAGTTILS I++TVDPST
Sbjct: 421 TPLLPLPKKAGKILVAGSHADDLGNQCGGWTITWQGQTGNDKTAGTTILSAIKSTVDPST 480
Query: 1823 QVVYSESPDSGVL-ADKYDYAIVVVGEPPYAETFGDNLNLTIPAPGPSVIQSVCGAAKCV 1999
+VV+SE+PDS + + KYDYAIVVVGEPPYAETFGDNLNLTIPAPGPSVIQ+VC + +CV
Sbjct: 481 EVVFSENPDSAAVDSGKYDYAIVVVGEPPYAETFGDNLNLTIPAPGPSVIQNVCKSVRCV 540
Query: 2000 VVLISGRPLVVEPYLGDMDALVAAWLPGSEGQGVADVLFGDYGFTGKLPRTWFKSVDQLP 2179
VVLISGRPLVVEPY+ MDA VAAWLPGSEGQGVADVLFGDYGF+GKL RTWFKS DQLP
Sbjct: 541 VVLISGRPLVVEPYISAMDAFVAAWLPGSEGQGVADVLFGDYGFSGKLARTWFKSADQLP 600
Query: 2180 MNVGDAHYDPLFPFGFGLTTKGTK 2251
MNVGD HYDPLFPFGFGLTT+ K
Sbjct: 601 MNVGDKHYDPLFPFGFGLTTEAKK 624
>sptr|Q8RWR5|Q8RWR5 Beta-D-glucan exohydrolase.
Length = 624
Score = 1077 bits (2786), Expect = 0.0
Identities = 529/624 (84%), Positives = 567/624 (90%), Gaps = 2/624 (0%)
Frame = +2
Query: 386 MASVHKATTLVLMFCLLALGRAEYLKYKDPKQPVAVRIKDLLGRMTLAEKIGQMTQIERE 565
M S+HK T ++LMFCL ALG A+YLKYKDPKQP+ VRIKDLLGRMTLAEKIGQMTQIERE
Sbjct: 1 MGSLHKTTFVLLMFCLAALGSADYLKYKDPKQPLGVRIKDLLGRMTLAEKIGQMTQIERE 60
Query: 566 NATADALAKYFIGSVLSGGGSVXXXXXXXXXXXXMVTEMQKGALSTRLGIPIIYGIDAVH 745
NATA+A++KYFIGSVLSGGGSV MV EMQKGALSTRLGIP+IYGIDAVH
Sbjct: 61 NATAEAMSKYFIGSVLSGGGSVPSPQASAAAWQSMVNEMQKGALSTRLGIPMIYGIDAVH 120
Query: 746 GHNNVYKATIFPHNVGLGATRDPDLVKRIGEATALEVRATGIPYAFAPCIAVCRDPRWGR 925
GHNNVYKATIFPHNVGLGATRDP L+KRIGEATALEVRATGIPYAFAPCIAVCRDPRWGR
Sbjct: 121 GHNNVYKATIFPHNVGLGATRDPMLIKRIGEATALEVRATGIPYAFAPCIAVCRDPRWGR 180
Query: 926 CYESYSEDPKVVQSLTSLISGLQGDAPADSAGRPYVGGSKKVAACAKHYVGDGGTHNGIN 1105
CYESYSEDPKVVQS+T+LISGLQGD PA S GRPYVGGSKKVAACAKHYVGDGGT GIN
Sbjct: 181 CYESYSEDPKVVQSMTTLISGLQGDVPAGSEGRPYVGGSKKVAACAKHYVGDGGTFMGIN 240
Query: 1106 ENNTIIDTHGLLSIHMPPYYNSIIRGVSTVMVSYSSWNGVKMHANHFLVTDFLKNKLKFR 1285
EN+TIID HGL++IHMP YYNSIIRGVSTVM SYSSWNG KMHANHFLVTDFLKNKLKFR
Sbjct: 241 ENDTIIDAHGLMTIHMPAYYNSIIRGVSTVMTSYSSWNGKKMHANHFLVTDFLKNKLKFR 300
Query: 1286 GFVISDWEGIDRITTPPHANYSYSIEAGVGAGIDMIMVPFRYTEFIDDLTTQVQNKVIPM 1465
GFVISDW+GIDRIT+PP NYSYS+EAGVGAGIDMIMVP+ YTEFIDDLT QV+N +IPM
Sbjct: 301 GFVISDWQGIDRITSPPGVNYSYSVEAGVGAGIDMIMVPYAYTEFIDDLTYQVKNNIIPM 360
Query: 1466 SRIDDAVYRILRVKFTMGLFENPYPDSSLAGELGKQEHRELAREAVRKSLVLLKNGKSSY 1645
SRIDDAVYRILRVKFTMGLFE+PY D SL GELGKQEHR+LAREAVRKSLVLLKNGKS+
Sbjct: 361 SRIDDAVYRILRVKFTMGLFESPYADPSLVGELGKQEHRDLAREAVRKSLVLLKNGKSAS 420
Query: 1646 APLLPLPKKAGKILVAGSHANDLGNQCGGWTITWQGSSGN-TTAGTTILSGIEATVDPST 1822
+PLLPLPKKAGKILVAGSHA+DLG QCGGWTITWQG +GN TAGTTILS I++TVDPST
Sbjct: 421 SPLLPLPKKAGKILVAGSHADDLGLQCGGWTITWQGQTGNDKTAGTTILSAIKSTVDPST 480
Query: 1823 QVVYSESPDSGVL-ADKYDYAIVVVGEPPYAETFGDNLNLTIPAPGPSVIQSVCGAAKCV 1999
+VV+SE+PDS + + KYDYAIVVVGE PYAETFGDNLNLTIPAPGPSVIQSVC +A CV
Sbjct: 481 EVVFSENPDSAAVDSGKYDYAIVVVGEQPYAETFGDNLNLTIPAPGPSVIQSVCKSANCV 540
Query: 2000 VVLISGRPLVVEPYLGDMDALVAAWLPGSEGQGVADVLFGDYGFTGKLPRTWFKSVDQLP 2179
VVLISGRPLVVEPY+G MDA VAAWLPGSEGQGVAD LFGDYGF+GKL RTWFKSVDQLP
Sbjct: 541 VVLISGRPLVVEPYIGAMDAFVAAWLPGSEGQGVADALFGDYGFSGKLARTWFKSVDQLP 600
Query: 2180 MNVGDAHYDPLFPFGFGLTTKGTK 2251
MNVGD HYDPLFPFGFGLTT+ K
Sbjct: 601 MNVGDKHYDPLFPFGFGLTTEANK 624
>sptr|Q7XAS3|Q7XAS3 Beta-D-glucosidase.
Length = 628
Score = 955 bits (2469), Expect = 0.0
Identities = 467/614 (76%), Positives = 524/614 (85%), Gaps = 3/614 (0%)
Frame = +2
Query: 416 VLMFCLLALGRAEYLKYKDPKQPVAVRIKDLLGRMTLAEKIGQMTQIERENATADALAKY 595
+L+ CL AL A Y+KYKDPKQP+ VRIKDL+ RMTLAEKIGQMTQIER AT DA+ Y
Sbjct: 12 LLLCCLAALTEATYVKYKDPKQPLGVRIKDLMRRMTLAEKIGQMTQIERTVATPDAMKNY 71
Query: 596 FIGSVLSGGGSVXXXXXXXXXXXXMVTEMQKGALSTRLGIPIIYGIDAVHGHNNVYKATI 775
FIGSVLSGGGSV MV MQK +LSTRLGIP+IYGIDAVHGHNNVYKATI
Sbjct: 72 FIGSVLSGGGSVPAQKATPETWIEMVNTMQKASLSTRLGIPMIYGIDAVHGHNNVYKATI 131
Query: 776 FPHNVGLGATRDPDLVKRIGEATALEVRATGIPYAFAPCIAVCRDPRWGRCYESYSEDPK 955
FPHN+GLG TRDP+LVKRIGEATALEVRATGIPY FAPCIAVCRDPRWGRCYESYSED K
Sbjct: 132 FPHNIGLGVTRDPNLVKRIGEATALEVRATGIPYVFAPCIAVCRDPRWGRCYESYSEDHK 191
Query: 956 VVQSLTSLISGLQGDAPADSA-GRPYVGGSKKVAACAKHYVGDGGTHNGINENNTIIDTH 1132
+VQ +T +I+GLQG P S G P+V G KVAACAKHYVGDGGT GINENNT+I +
Sbjct: 192 IVQMMTEIITGLQGGLPVHSKKGVPFVAGKNKVAACAKHYVGDGGTTKGINENNTVISWN 251
Query: 1133 GLLSIHMPPYYNSIIRGVSTVMVSYSSWNGVKMHANHFLVTDFLKNKLKFRGFVISDWEG 1312
GLL IHMP Y+NSI +GV+T+M SYSSWNG KMHANH LVTDFLKNKLKFRGFVISDW+G
Sbjct: 252 GLLGIHMPAYFNSIAKGVATIMTSYSSWNGKKMHANHDLVTDFLKNKLKFRGFVISDWQG 311
Query: 1313 IDRITTPPHANYSYSIEAGVGAGIDMIMVPFRYTEFIDDLTTQVQNKVIPMSRIDDAVYR 1492
+DRIT+PPHANYSYS+EAGVGAGIDM+MVP+ +TEFIDDLT QV+N +IPMSRIDDAV R
Sbjct: 312 LDRITSPPHANYSYSVEAGVGAGIDMVMVPYNFTEFIDDLTYQVKNNIIPMSRIDDAVKR 371
Query: 1493 ILRVKFTMGLFENPYPDSSLAGELGKQEHRELAREAVRKSLVLLKNGKSSYAPLLPLPKK 1672
ILRVKF MGLFENP D+SL +LG QEHRELAREAVRKSLVLLKNG+S+ PLLPLPKK
Sbjct: 372 ILRVKFVMGLFENPMADNSLVNQLGSQEHRELAREAVRKSLVLLKNGESADKPLLPLPKK 431
Query: 1673 AGKILVAGSHANDLGNQCGGWTITWQGSSGN-TTAGTTILSGIEATVDPSTQVVYSESPD 1849
A KILVAG+HA++LG QCGGWTITWQG GN T GTTIL ++ TVD STQVVYSE+PD
Sbjct: 432 ATKILVAGTHADNLGYQCGGWTITWQGLGGNDLTTGTTILQAVKNTVDSSTQVVYSENPD 491
Query: 1850 SG-VLADKYDYAIVVVGEPPYAETFGDNLNLTIPAPGPSVIQSVCGAAKCVVVLISGRPL 2026
+G V + ++ YAIVVVGEPPYAET+GD+LNLTI PGP I +VCG+ KCVVV+ISGRP+
Sbjct: 492 AGFVKSGEFSYAIVVVGEPPYAETYGDSLNLTISEPGPMTIYNVCGSVKCVVVVISGRPV 551
Query: 2027 VVEPYLGDMDALVAAWLPGSEGQGVADVLFGDYGFTGKLPRTWFKSVDQLPMNVGDAHYD 2206
VV+P++ +DALVAAWLPG+EGQGV+DVLFGDYGFTGKL RTWFK+VDQLPMNVGD HYD
Sbjct: 552 VVQPFVSSVDALVAAWLPGTEGQGVSDVLFGDYGFTGKLARTWFKTVDQLPMNVGDPHYD 611
Query: 2207 PLFPFGFGLTTKGT 2248
PLFPFGFGLTTK T
Sbjct: 612 PLFPFGFGLTTKPT 625
>sptr|O82074|O82074 Beta-D-glucosidase precursor (EC 3.2.1.21).
Length = 654
Score = 928 bits (2398), Expect = 0.0
Identities = 454/612 (74%), Positives = 518/612 (84%), Gaps = 3/612 (0%)
Frame = +2
Query: 416 VLMFCLLALGRAEYLKYKDPKQPVAVRIKDLLGRMTLAEKIGQMTQIERENATADALAKY 595
+L+ CL A AEY++YKDPK+P+ VRIKDL+ RMTLAEKIGQMTQIER+ AT D ++KY
Sbjct: 13 LLLSCLSAFTEAEYMRYKDPKKPLNVRIKDLMSRMTLAEKIGQMTQIERKEATPDVISKY 72
Query: 596 FIGSVLSGGGSVXXXXXXXXXXXXMVTEMQKGALSTRLGIPIIYGIDAVHGHNNVYKATI 775
FIGSVLSGGGSV +V MQK ALSTRLGIP+IYGIDAVHGHNNVY ATI
Sbjct: 73 FIGSVLSGGGSVPAPKASPEAWVDLVNGMQKAALSTRLGIPMIYGIDAVHGHNNVYNATI 132
Query: 776 FPHNVGLGATRDPDLVKRIGEATALEVRATGIPYAFAPCIAVCRDPRWGRCYESYSEDPK 955
FPHNVGLG TRDP L+KRIGEATALE RATGIPYAFAPCIAVCRDPRWGRCYESYSED
Sbjct: 133 FPHNVGLGVTRDPALIKRIGEATALECRATGIPYAFAPCIAVCRDPRWGRCYESYSEDHT 192
Query: 956 VVQSLTSLISGLQGDAPAD-SAGRPYVGGSKKVAACAKHYVGDGGTHNGINENNTIIDTH 1132
+VQ++T +I GLQGD P D G P+VGG KVAACAKH+VGDGGT GI+ENNT+ID+
Sbjct: 193 IVQAMTEIIPGLQGDVPPDVKKGVPFVGGKTKVAACAKHFVGDGGTTKGIDENNTVIDSR 252
Query: 1133 GLLSIHMPPYYNSIIRGVSTVMVSYSSWNGVKMHANHFLVTDFLKNKLKFRGFVISDWEG 1312
GL SIHMP Y++SI +GV+TVMVSYSSWNG++MHAN LVT +LKNKLKFRGFVISDWEG
Sbjct: 253 GLFSIHMPAYHDSIKKGVATVMVSYSSWNGLRMHANRDLVTGYLKNKLKFRGFVISDWEG 312
Query: 1313 IDRITTPPHANYSYSIEAGVGAGIDMIMVPFRYTEFIDDLTTQVQNKVIPMSRIDDAVYR 1492
IDRIT PP NYSYS+EAGVGAGIDMIMVP +T+F+++LT+QV+ +IPMSRIDDAV R
Sbjct: 313 IDRITDPPGRNYSYSVEAGVGAGIDMIMVPEDFTKFLNELTSQVKKNIIPMSRIDDAVKR 372
Query: 1493 ILRVKFTMGLFENPYPDSSLAGELGKQEHRELAREAVRKSLVLLKNGKSSYAPLLPLPKK 1672
ILRVKF MGLFE+P D SLA +LG QEHR+LAREAVRKSLVLLKNG+S+ P +PLPK
Sbjct: 373 ILRVKFVMGLFESPLADYSLANQLGSQEHRDLAREAVRKSLVLLKNGESADKPFVPLPKN 432
Query: 1673 AGKILVAGSHANDLGNQCGGWTITWQGSSGN-TTAGTTILSGIEATVDPSTQVVYSESPD 1849
A KILVAGSHA++LG QCGGWTI WQG +GN T GTTIL+ I+ TVDP+TQV+Y+E+PD
Sbjct: 433 AKKILVAGSHADNLGRQCGGWTIEWQGVNGNDLTTGTTILNAIKKTVDPTTQVIYNENPD 492
Query: 1850 SG-VLADKYDYAIVVVGEPPYAETFGDNLNLTIPAPGPSVIQSVCGAAKCVVVLISGRPL 2026
S V + +DYAIVVVGEPPYAE GD+ NLTIP PGP+ I SVCGA KCVVV+ISGRP+
Sbjct: 493 SNYVKTNSFDYAIVVVGEPPYAEMQGDSFNLTIPEPGPTTISSVCGAVKCVVVVISGRPV 552
Query: 2027 VVEPYLGDMDALVAAWLPGSEGQGVADVLFGDYGFTGKLPRTWFKSVDQLPMNVGDAHYD 2206
V++PY+ MDALVAAWLPG+EGQGV DVLFGDYGFTGKL RTWFK+VDQLPMNVGD HYD
Sbjct: 553 VLQPYVSYMDALVAAWLPGTEGQGVTDVLFGDYGFTGKLARTWFKTVDQLPMNVGDKHYD 612
Query: 2207 PLFPFGFGLTTK 2242
PLFPFGFGLTTK
Sbjct: 613 PLFPFGFGLTTK 624
>sptr|Q8W112|Q8W112 Beta-D-glucan exohydrolase-like protein.
Length = 624
Score = 924 bits (2389), Expect = 0.0
Identities = 447/624 (71%), Positives = 520/624 (83%), Gaps = 2/624 (0%)
Frame = +2
Query: 386 MASVHKATTLVLMFCLLALGRAEYLKYKDPKQPVAVRIKDLLGRMTLAEKIGQMTQIERE 565
M ++ K L+L+ C++A LKYKDPKQP+ RI+DL+ RMTL EKIGQM QIER
Sbjct: 1 MGTLSKVLCLMLLCCIVAAAEGT-LKYKDPKQPLGARIRDLMNRMTLQEKIGQMVQIERS 59
Query: 566 NATADALAKYFIGSVLSGGGSVXXXXXXXXXXXXMVTEMQKGALSTRLGIPIIYGIDAVH 745
AT + + KYFIGSVLSGGGSV MV E+QK +LSTRLGIP+IYGIDAVH
Sbjct: 60 VATPEVMKKYFIGSVLSGGGSVPSEKATPETWVNMVNEIQKASLSTRLGIPMIYGIDAVH 119
Query: 746 GHNNVYKATIFPHNVGLGATRDPDLVKRIGEATALEVRATGIPYAFAPCIAVCRDPRWGR 925
GHNNVY ATIFPHNVGLG TRDP+LVKRIG ATALEVRATGIPYAFAPCIAVCRDPRWGR
Sbjct: 120 GHNNVYGATIFPHNVGLGVTRDPNLVKRIGAATALEVRATGIPYAFAPCIAVCRDPRWGR 179
Query: 926 CYESYSEDPKVVQSLTSLISGLQGDAPADSAGRPYVGGSKKVAACAKHYVGDGGTHNGIN 1105
CYESYSED ++VQ +T +I GLQGD P G P+VGG KVAACAKH+VGDGGT GI+
Sbjct: 180 CYESYSEDYRIVQQMTEIIPGLQGDLPTKRKGVPFVGGKTKVAACAKHFVGDGGTVRGID 239
Query: 1106 ENNTIIDTHGLLSIHMPPYYNSIIRGVSTVMVSYSSWNGVKMHANHFLVTDFLKNKLKFR 1285
ENNT+ID+ GL IHMP YYN++ +GV+T+MVSYS+WNG++MHAN LVT FLKNKLKFR
Sbjct: 240 ENNTVIDSKGLFGIHMPGYYNAVNKGVATIMVSYSAWNGLRMHANKELVTGFLKNKLKFR 299
Query: 1286 GFVISDWEGIDRITTPPHANYSYSIEAGVGAGIDMIMVPFRYTEFIDDLTTQVQNKVIPM 1465
GFVISDW+GIDRITTPPH NYSYS+ AG+ AGIDMIMVP+ YTEFID++++Q+Q K+IP+
Sbjct: 300 GFVISDWQGIDRITTPPHLNYSYSVYAGISAGIDMIMVPYNYTEFIDEISSQIQKKLIPI 359
Query: 1466 SRIDDAVYRILRVKFTMGLFENPYPDSSLAGELGKQEHRELAREAVRKSLVLLKNGKSSY 1645
SRIDDA+ RILRVKFTMGLFE P D S A +LG +EHRELAREAVRKSLVLLKNGK+
Sbjct: 360 SRIDDALKRILRVKFTMGLFEEPLADLSFANQLGSKEHRELAREAVRKSLVLLKNGKTGA 419
Query: 1646 APLLPLPKKAGKILVAGSHANDLGNQCGGWTITWQGSSGNT-TAGTTILSGIEATVDPST 1822
PLLPLPKK+GKILVAG+HA++LG QCGGWTITWQG +GN T GTTIL+ ++ TV P+T
Sbjct: 420 KPLLPLPKKSGKILVAGAHADNLGYQCGGWTITWQGLNGNDHTVGTTILAAVKNTVAPTT 479
Query: 1823 QVVYSESPDSG-VLADKYDYAIVVVGEPPYAETFGDNLNLTIPAPGPSVIQSVCGAAKCV 1999
QVVYS++PD+ V + K+DYAIVVVGEPPYAE FGD NLTI PGPS+I +VCG+ KCV
Sbjct: 480 QVVYSQNPDANFVKSGKFDYAIVVVGEPPYAEMFGDTTNLTISDPGPSIIGNVCGSVKCV 539
Query: 2000 VVLISGRPLVVEPYLGDMDALVAAWLPGSEGQGVADVLFGDYGFTGKLPRTWFKSVDQLP 2179
VV++SGRP+V++PY+ +DALVAAWLPG+EGQGVAD LFGDYGFTGKL RTWFKSV QLP
Sbjct: 540 VVVVSGRPVVIQPYVSTIDALVAAWLPGTEGQGVADALFGDYGFTGKLARTWFKSVKQLP 599
Query: 2180 MNVGDAHYDPLFPFGFGLTTKGTK 2251
MNVGD HYDPL+PFGFGLTTK K
Sbjct: 600 MNVGDRHYDPLYPFGFGLTTKPYK 623
>sptr|O82151|O82151 Beta-D-glucan exohydrolase.
Length = 628
Score = 914 bits (2361), Expect = 0.0
Identities = 453/615 (73%), Positives = 515/615 (83%), Gaps = 4/615 (0%)
Frame = +2
Query: 419 LMFCLLAL-GRAEYLKYKDPKQPVAVRIKDLLGRMTLAEKIGQMTQIERENATADALAKY 595
++ CL A+ EY+KYKDPKQPV RIKDL+ RMTL EKIGQMTQIER+ ATAD + +
Sbjct: 12 VVLCLWAVVAEGEYVKYKDPKQPVGARIKDLMKRMTLEEKIGQMTQIERKVATADVMKQN 71
Query: 596 FIGSVLSGGGSVXXXXXXXXXXXXMVTEMQKGALSTRLGIPIIYGIDAVHGHNNVYKATI 775
FIGSVLSGGGSV MV E+QKG+LSTRLGIP+IYGIDAVHGHNNVY ATI
Sbjct: 72 FIGSVLSGGGSVPAPKASAQVWTNMVDEIQKGSLSTRLGIPMIYGIDAVHGHNNVYGATI 131
Query: 776 FPHNVGLGATRDPDLVKRIGEATALEVRATGIPYAFAPCIAVCRDPRWGRCYESYSEDPK 955
FPHNVGLG TRDPDLVKRIG ATALEVRATGIPYAFAPCIAVCR+PRWGRCYESYSED +
Sbjct: 132 FPHNVGLGVTRDPDLVKRIGAATALEVRATGIPYAFAPCIAVCRNPRWGRCYESYSEDHR 191
Query: 956 VVQSLTSLISGLQGDAPADSA-GRPYVGGSKKVAACAKHYVGDGGTHNGINENNTIIDTH 1132
+V+S+T +I GLQGD PA S G PYVGG KVAACAKH+VGDGGT +G++E+NT+I ++
Sbjct: 192 IVRSMTEIIPGLQGDLPAKSKNGVPYVGGKTKVAACAKHFVGDGGTLHGVDESNTVISSN 251
Query: 1133 GLLSIHMPPYYNSIIRGVSTVMVSYSSWNGVKMHANHFLVTDFLKNKLKFRGFVISDWEG 1312
L SIHMP YY+S+ +GV+TVMVSYSSWNG KMHAN LVT FLK+KLKFRGFVISDW+G
Sbjct: 252 SLFSIHMPAYYDSLRKGVATVMVSYSSWNGRKMHANRDLVTGFLKDKLKFRGFVISDWQG 311
Query: 1313 IDRITTPPHANYSYSIEAGVGAGIDMIMVPFRYTEFIDDLTTQVQNKVIPMSRIDDAVYR 1492
IDRIT PPHANYSYS++AG+ AGIDMIMVP Y EFID LT+QV+ +IPMSRIDDAV R
Sbjct: 312 IDRITDPPHANYSYSVQAGIMAGIDMIMVPENYREFIDTLTSQVKANIIPMSRIDDAVKR 371
Query: 1493 ILRVKFTMGLFENPYPDSSLAGELGKQEHRELAREAVRKSLVLLKNGKSSYAPLLPLPKK 1672
ILRVKF MGLFENP D SLA +LG QEHRELAREAVRKSLVLLKNGK+ PLLPLPKK
Sbjct: 372 ILRVKFVMGLFENPMSDPSLANQLGSQEHRELAREAVRKSLVLLKNGKTPSQPLLPLPKK 431
Query: 1673 AGKILVAGSHANDLGNQCGGWTITWQGSSGN-TTAGTTILSGIEATVDPSTQVVYSESPD 1849
A KILVAG+HA++LG QCGGWTI WQG +GN T GTTIL+ I+ TVDPSTQVVY ++PD
Sbjct: 432 APKILVAGTHADNLGYQCGGWTIEWQGVAGNDLTIGTTILTAIKKTVDPSTQVVYQQNPD 491
Query: 1850 SG-VLADKYDYAIVVVGEPPYAETFGDNLNLTIPAPGPSVIQSVCGAAKCVVVLISGRPL 2026
+ V ++K+ YAIVVVGE PYAE FGD+ NLTI PGPS I ++CG+ KCVVV++SGRP+
Sbjct: 492 ANFVKSNKFSYAIVVVGEVPYAEMFGDSSNLTIAEPGPSTISNICGSVKCVVVVVSGRPV 551
Query: 2027 VVEPYLGDMDALVAAWLPGSEGQGVADVLFGDYGFTGKLPRTWFKSVDQLPMNVGDAHYD 2206
V+EPY+ MDALVAAWLPG+EGQGVAD LFGDYGFTGKL RTWFK VDQLPMN DAH D
Sbjct: 552 VLEPYVSKMDALVAAWLPGTEGQGVADALFGDYGFTGKLARTWFKRVDQLPMNFDDAHVD 611
Query: 2207 PLFPFGFGLTTKGTK 2251
PLFPFGFG+TTK K
Sbjct: 612 PLFPFGFGITTKPVK 626
>sptrnew|BAD07748|BAD07748 Putative beta-D-glucan exohydrolase.
Length = 648
Score = 907 bits (2343), Expect = 0.0
Identities = 442/608 (72%), Positives = 508/608 (83%), Gaps = 8/608 (1%)
Frame = +2
Query: 443 GRAEYLKYKDPKQPVAVRIKDLLGRMTLAEKIGQMTQIERENATADALAKYFIGSVLSGG 622
G E KYKD KQP+ RI DLL RMTLAEKIGQM+QIERENAT D + YFIGSVLSGG
Sbjct: 38 GGGECPKYKDSKQPLNKRIDDLLRRMTLAEKIGQMSQIERENATFDVMRNYFIGSVLSGG 97
Query: 623 GSVXXXXXXXXXXXXMVTEMQKGALSTRLGIPIIYGIDAVHGHNNVYKATIFPHNVGLGA 802
GSV MV EMQ+GA++TRLGIP+IYGIDAVHGH NVYKATIFPHNVGLG
Sbjct: 98 GSVPAAQASPAAWVSMVNEMQRGAMATRLGIPMIYGIDAVHGHGNVYKATIFPHNVGLGC 157
Query: 803 TRDPDLVKRIGEATALEVRATGIPYAFAPCIAVCRDPRWGRCYESYSEDPKVVQSLTSLI 982
TRDPDL KRIG A A EVRATGIPY FAPC+AVCRDPRWGRCYES+SEDP+VVQ ++S+I
Sbjct: 158 TRDPDLAKRIGAAVAAEVRATGIPYVFAPCVAVCRDPRWGRCYESFSEDPRVVQRMSSII 217
Query: 983 SGLQGDAPADSA-GRPYV-GGSKKVAACAKHYVGDGGTHNGINENNTIIDTHGLLSIHMP 1156
SG QG+ P G P+V GG VAAC+KHYVGDGGT G+NENNT+ L+++HMP
Sbjct: 218 SGFQGEIPPGGRRGVPFVSGGRPSVAACSKHYVGDGGTTRGMNENNTVATLRELMTVHMP 277
Query: 1157 PYYNSIIRGVSTVMVSYSSWNGVKMHANHFLVTDFLKNKLKFRGFVISDWEGIDRITTPP 1336
PYY+++ +GVSTVMVS+SSWNGVKMHANHFL+TDFLK+KL+FRGFVISDW+G+DRITTP
Sbjct: 278 PYYSAVAQGVSTVMVSFSSWNGVKMHANHFLITDFLKSKLRFRGFVISDWQGLDRITTPA 337
Query: 1337 HANYSYSIEAGVGAGIDMIMVPFRYTEFIDDLTTQVQNKVIPMSRIDDAVYRILRVKFTM 1516
HA+Y SI+ G+ AGIDM+M+PF YTEFIDDL V+N IPMSRIDDAV RILRVKFTM
Sbjct: 338 HADYMLSIKLGIMAGIDMVMIPFTYTEFIDDLAALVKNGTIPMSRIDDAVRRILRVKFTM 397
Query: 1517 GLFENPYPDSSLAGELGKQEHRELAREAVRKSLVLLKNGKSSYAPLLPLPKKAGKILVAG 1696
GLFE PY D SLAGELGKQEHR+LAR+AVRKSLVLLKNGK APLLPLPK+A ILVAG
Sbjct: 398 GLFERPYADLSLAGELGKQEHRDLARDAVRKSLVLLKNGKPGDAPLLPLPKRARSILVAG 457
Query: 1697 SHANDLGNQCGGWTITWQGSSGN--TTAGTTILSGIEATVDPSTQVVYSESPDSGVL--- 1861
+HA+DLG+QCGGWTITWQG +GN T GTTIL GI VD +T+VV++E+PD+G +
Sbjct: 458 AHADDLGSQCGGWTITWQGLAGNDLTAGGTTILDGIRRAVDAATEVVFAEAPDAGFMRRN 517
Query: 1862 ADKYDYAIVVVGEPPYAETFGDNLNLTIPAPGPSVIQSVC-GAAKCVVVLISGRPLVVEP 2038
A ++D A+VVVGEPPYAET GDNLNLTIPAPGPSVIQ+VC G +CVVV++SGRPLV+EP
Sbjct: 518 AGRFDAAVVVVGEPPYAETLGDNLNLTIPAPGPSVIQNVCGGGVRCVVVVVSGRPLVIEP 577
Query: 2039 YLGDMDALVAAWLPGSEGQGVADVLFGDYGFTGKLPRTWFKSVDQLPMNVGDAHYDPLFP 2218
Y+ +DALVAAWLPG+EGQGV+DVLFGDY FTGKL RTWF+SV+QLPMNVGD HYDPLFP
Sbjct: 578 YMDAIDALVAAWLPGTEGQGVSDVLFGDYEFTGKLARTWFRSVEQLPMNVGDEHYDPLFP 637
Query: 2219 FGFGLTTK 2242
FGFGL T+
Sbjct: 638 FGFGLETR 645
>sptr|Q8W2Z2|Q8W2Z2 Putative exohydrolase.
Length = 677
Score = 903 bits (2333), Expect = 0.0
Identities = 446/634 (70%), Positives = 517/634 (81%), Gaps = 2/634 (0%)
Frame = +2
Query: 347 RWFEVIGRTTQAKMASVHKATTLVLMFCLLALGRAEYLKYKDPKQPVAVRIKDLLGRMTL 526
R ++ R A +A+ A L+L + A G A+Y+ YKD +PV R+ DLL RMTL
Sbjct: 42 RLLPLLRRQRMAVLAAPVVAAVLLLTWA--AYGEAQYVLYKDATKPVEARVTDLLARMTL 99
Query: 527 AEKIGQMTQIERENATADALAKYFIGSVLSGGGSVXXXXXXXXXXXXMVTEMQKGALSTR 706
AEKIGQMTQIER+ A+ L YFIGS+LSGGGSV MV++ QKG+LSTR
Sbjct: 100 AEKIGQMTQIERQVASPQVLKDYFIGSLLSGGGSVPRKQATAAEWVSMVSDFQKGSLSTR 159
Query: 707 LGIPIIYGIDAVHGHNNVYKATIFPHNVGLGATRDPDLVKRIGEATALEVRATGIPYAFA 886
LGIP+IYGIDAVHGHNNVY ATIFPHNV LGATRDP+LVKRIG ATALEVRATGI YAFA
Sbjct: 160 LGIPMIYGIDAVHGHNNVYGATIFPHNVALGATRDPNLVKRIGAATALEVRATGIQYAFA 219
Query: 887 PCIAVCRDPRWGRCYESYSEDPKVVQSLTSLISGLQGDAPAD-SAGRPYVGGSKKVAACA 1063
PCIAVCRDPRWGRCYESYSED ++VQ++T LI GLQGD PA+ ++G PYV G VAACA
Sbjct: 220 PCIAVCRDPRWGRCYESYSEDHRIVQAMTELIPGLQGDVPANFTSGMPYVAGKNNVAACA 279
Query: 1064 KHYVGDGGTHNGINENNTIIDTHGLLSIHMPPYYNSIIRGVSTVMVSYSSWNGVKMHANH 1243
KH+VGDGGT NG+NE+NTIID GL++IHMP Y N++ +GVSTVM+SYSSWNG+KMHANH
Sbjct: 280 KHFVGDGGTQNGVNEDNTIIDRRGLMTIHMPAYLNALQKGVSTVMISYSSWNGIKMHANH 339
Query: 1244 FLVTDFLKNKLKFRGFVISDWEGIDRITTPPHANYSYSIEAGVGAGIDMIMVPFRYTEFI 1423
LVT +LK++L F+GF ISDWEGIDRITTP +NYSYS++AGV AGIDMIMVP Y FI
Sbjct: 340 DLVTRYLKDRLNFKGFTISDWEGIDRITTPAGSNYSYSVQAGVLAGIDMIMVPNNYQSFI 399
Query: 1424 DDLTTQVQNKVIPMSRIDDAVYRILRVKFTMGLFENPYPDSSLAGELGKQEHRELAREAV 1603
LT+ V N +IPMSRIDDAV RILRVKFTMGLFENP PDSS+A +LGK+EHR+LAREAV
Sbjct: 400 SILTSHVNNGIIPMSRIDDAVTRILRVKFTMGLFENPMPDSSMADQLGKKEHRDLAREAV 459
Query: 1604 RKSLVLLKNGKSSYAPLLPLPKKAGKILVAGSHANDLGNQCGGWTITWQGSSGNTTAGTT 1783
RKSLVLLKNGK+S P+LPL KKA KILVAGSHA++LG QCGGWTI WQG +G T G T
Sbjct: 460 RKSLVLLKNGKTSDKPMLPLSKKAPKILVAGSHADNLGYQCGGWTIEWQGDTGRITVGMT 519
Query: 1784 ILSGIEATVDPSTQVVYSESPDSGVLAD-KYDYAIVVVGEPPYAETFGDNLNLTIPAPGP 1960
IL ++A VDPST VV++E+PD+ + + + YAIVVVGE PY ET GD+LNLTIP PGP
Sbjct: 520 ILDAVKAAVDPSTTVVFAENPDADFVKNGGFSYAIVVVGEHPYTETKGDSLNLTIPDPGP 579
Query: 1961 SVIQSVCGAAKCVVVLISGRPLVVEPYLGDMDALVAAWLPGSEGQGVADVLFGDYGFTGK 2140
S + +VCGAA+C VLISGRP+VV+P+LG MDALVAAWLPG+EGQGV DVLFGDYGFTGK
Sbjct: 580 STVATVCGAAQCATVLISGRPVVVQPFLGAMDALVAAWLPGTEGQGVTDVLFGDYGFTGK 639
Query: 2141 LPRTWFKSVDQLPMNVGDAHYDPLFPFGFGLTTK 2242
LPRTWFKSVDQLPMN GDAHYDPLFP GFGLTT+
Sbjct: 640 LPRTWFKSVDQLPMNYGDAHYDPLFPLGFGLTTQ 673
>sptr|Q94ED2|Q94ED2 Putative beta-glucosidase.
Length = 663
Score = 895 bits (2314), Expect = 0.0
Identities = 442/615 (71%), Positives = 503/615 (81%), Gaps = 2/615 (0%)
Frame = +2
Query: 413 LVLMFCLLALGRAEYLKYKDPKQPVAVRIKDLLGRMTLAEKIGQMTQIERENATADALAK 592
++L+ + G A Y+KY DPKQP RIKDL+ RMTLAEKIGQMTQIER A+AD +
Sbjct: 11 VLLLLWFTSTGDAAYMKYLDPKQPTNTRIKDLISRMTLAEKIGQMTQIERGVASADVMKN 70
Query: 593 YFIGSVLSGGGSVXXXXXXXXXXXXMVTEMQKGALSTRLGIPIIYGIDAVHGHNNVYKAT 772
YFIGSVLSGGGSV MV E QKGALSTRLGIP+IYGIDAVHG+NNVY AT
Sbjct: 71 YFIGSVLSGGGSVPAPQATPAVWVNMVNEFQKGALSTRLGIPMIYGIDAVHGNNNVYNAT 130
Query: 773 IFPHNVGLGATRDPDLVKRIGEATALEVRATGIPYAFAPCIAVCRDPRWGRCYESYSEDP 952
+FPHN+GLGATRDPDLV+RIGEATALEVRATGIPY FAPCIAVCRDPRWGRCYESYSED
Sbjct: 131 LFPHNIGLGATRDPDLVRRIGEATALEVRATGIPYTFAPCIAVCRDPRWGRCYESYSEDH 190
Query: 953 KVVQSLTSLISGLQGDAPAD-SAGRPYVGGSKKVAACAKHYVGDGGTHNGINENNTIIDT 1129
+VVQ +T +I GLQGD P + + G PY+ G KVAACAKH+VGDGGTHNGINENNTI D
Sbjct: 191 RVVQQMTDIILGLQGDIPINHTKGVPYIAGKDKVAACAKHFVGDGGTHNGINENNTITDE 250
Query: 1130 HGLLSIHMPPYYNSIIRGVSTVMVSYSSWNGVKMHANHFLVTDFLKNKLKFRGFVISDWE 1309
HGLL IHMPPYY+SII+GV+TVMVSYSS NGVKMHANH LVT +LK+KL FRGFVISDW
Sbjct: 251 HGLLGIHMPPYYDSIIKGVATVMVSYSSLNGVKMHANHDLVTGYLKSKLHFRGFVISDWL 310
Query: 1310 GIDRITTPPHANYSYSIEAGVGAGIDMIMVPFRYTEFIDDLTTQVQNKVIPMSRIDDAVY 1489
GIDRIT+PP ANY+YS++AG+ AGIDM+MVPF YT++IDD+T+ V+ +I MSRIDDAV
Sbjct: 311 GIDRITSPPDANYTYSVQAGINAGIDMVMVPFNYTQYIDDVTSLVKKGIINMSRIDDAVR 370
Query: 1490 RILRVKFTMGLFENPYPDSSLAGELGKQEHRELAREAVRKSLVLLKNGKSSYAPLLPLPK 1669
RILRVKF MGLFENP D S A +LGK+EHR+LAREAVRKSLVLLKNG S LPLPK
Sbjct: 371 RILRVKFIMGLFENPLADLSFADQLGKKEHRDLAREAVRKSLVLLKNGNSPNQQFLPLPK 430
Query: 1670 KAGKILVAGSHANDLGNQCGGWTITWQGSSGNTTAGTTILSGIEATVDPSTQVVYSESPD 1849
KA ILVAGSHA++LG QCGGW+I W G SG+ T GTTIL I++TV ST VVYSE+PD
Sbjct: 431 KARSILVAGSHASNLGYQCGGWSIEWIGGSGDITVGTTILEAIKSTVADSTHVVYSENPD 490
Query: 1850 SGVLADK-YDYAIVVVGEPPYAETFGDNLNLTIPAPGPSVIQSVCGAAKCVVVLISGRPL 2026
+ + + +AIVVVGE YAET GD+ LTI PG I++VC AKC VV+ISGRP+
Sbjct: 491 ESFMKNNDFSFAIVVVGERTYAETTGDDPELTILDPGTDTIRTVCSTAKCAVVIISGRPV 550
Query: 2027 VVEPYLGDMDALVAAWLPGSEGQGVADVLFGDYGFTGKLPRTWFKSVDQLPMNVGDAHYD 2206
V+EPYL M+ALVAAWLPG+EGQGVADVLFGDYGFTGKLPRTWFKSVDQLPMNVGD HYD
Sbjct: 551 VIEPYLPMMEALVAAWLPGTEGQGVADVLFGDYGFTGKLPRTWFKSVDQLPMNVGDLHYD 610
Query: 2207 PLFPFGFGLTTKGTK 2251
PLFPFGFGLT ++
Sbjct: 611 PLFPFGFGLTINSSQ 625
>sptr|Q8W2Z9|Q8W2Z9 Unnamed protein product.
Length = 644
Score = 890 bits (2301), Expect = 0.0
Identities = 430/603 (71%), Positives = 497/603 (82%), Gaps = 6/603 (0%)
Frame = +2
Query: 452 EYLKYKDPKQPVAVRIKDLLGRMTLAEKIGQMTQIERENATADALAKYFIGSVLSGGGSV 631
EY+KYKDPK+P+ R+ DLL RMTLAEKIGQM+QIER NAT+ + KYF+GSVLSGGGSV
Sbjct: 36 EYVKYKDPKKPIGERVDDLLSRMTLAEKIGQMSQIERANATSAVIEKYFVGSVLSGGGSV 95
Query: 632 XXXXXXXXXXXXMVTEMQKGALSTRLGIPIIYGIDAVHGHNNVYKATIFPHNVGLGATRD 811
MV +MQK AL TRLGIPIIYGIDAVHGHNNV+ ATIFPHNVGLGATRD
Sbjct: 96 PSEKATAKEWQQMVAKMQKAALKTRLGIPIIYGIDAVHGHNNVHNATIFPHNVGLGATRD 155
Query: 812 PDLVKRIGEATALEVRATGIPYAFAPCIAVCRDPRWGRCYESYSEDPKVVQSLTS-LISG 988
P LVKRIG++TA E RATGIPY FAPC+AVCRDPRWGRCYESYSED K+VQ +TS ++ G
Sbjct: 156 PKLVKRIGQSTAHEARATGIPYTFAPCVAVCRDPRWGRCYESYSEDTKLVQLMTSAMVPG 215
Query: 989 LQGDAPAD-SAGRPYVGGSKKVAACAKHYVGDGGTHNGINENNTIIDTHGLLSIHMPPYY 1165
LQGDAPA G P+V G VA CAKH+VGDGGT +GINENNT++ H L+ IHMPPY
Sbjct: 216 LQGDAPARYPKGTPFVAGGMNVAGCAKHFVGDGGTRDGINENNTVLSFHDLMRIHMPPYD 275
Query: 1166 NSIIRGVSTVMVSYSSWNGVKMHANHFLVTDFLKNKLKFRGFVISDWEGIDRITTPPHAN 1345
+++I+GV++VM+SYSSWNGVKMH N FL+TD LKNKLKFRGFVI+DW+ +DRITTPPH +
Sbjct: 276 DAVIKGVASVMISYSSWNGVKMHENRFLITDILKNKLKFRGFVITDWQAVDRITTPPHKH 335
Query: 1346 YSYSIEAGVGAGIDMIMVPFRYTEFIDDLTTQVQNKVIPMSRIDDAVYRILRVKFTMGLF 1525
Y +SI+ + AGIDM+M+P+ Y EF+ DLTTQV N I + RI+DAV RILRVKF MGLF
Sbjct: 336 YYHSIQETIHAGIDMVMIPYDYPEFVADLTTQVSNGSIKLDRINDAVSRILRVKFAMGLF 395
Query: 1526 ENPYPDSSLAGELGKQEHRELAREAVRKSLVLLKNGKSSYAPLLPLPKKAGKILVAGSHA 1705
ENP PD LAGELG +EHR++AREAVR+SLVLLKNGK P+LPL KKA KILVAGSHA
Sbjct: 396 ENPLPDPRLAGELGDKEHRQIAREAVRRSLVLLKNGKHGEKPVLPLSKKADKILVAGSHA 455
Query: 1706 NDLGNQCGGWTITWQGSSG-NTTAGTTILSGIEATVDPSTQVVYSESPDSGVLAD---KY 1873
++LG QCGGWT++WQG G N TAGTTIL I+A VD ST + Y+E PD +A+ +Y
Sbjct: 456 HNLGFQCGGWTVSWQGQGGNNVTAGTTILEAIKAAVDESTVIDYTEHPDKSSIAESAKEY 515
Query: 1874 DYAIVVVGEPPYAETFGDNLNLTIPAPGPSVIQSVCGAAKCVVVLISGRPLVVEPYLGDM 2053
DYA+VVVGE PYAET GDNLNLTIP+PGP VI+ VCG KCVVVL+SGRPLVVEPY+G M
Sbjct: 516 DYAVVVVGEEPYAETEGDNLNLTIPSPGPKVIKDVCGLVKCVVVLVSGRPLVVEPYIGAM 575
Query: 2054 DALVAAWLPGSEGQGVADVLFGDYGFTGKLPRTWFKSVDQLPMNVGDAHYDPLFPFGFGL 2233
DA VAAWLPG+EG GVADVLFGD+GFTGKLPRTWFKSVDQLPMN GD HY+PLFPFGFGL
Sbjct: 576 DAFVAAWLPGTEGHGVADVLFGDHGFTGKLPRTWFKSVDQLPMNFGDKHYNPLFPFGFGL 635
Query: 2234 TTK 2242
TTK
Sbjct: 636 TTK 638
>sptr|Q9XE93|Q9XE93 Exhydrolase II.
Length = 634
Score = 887 bits (2291), Expect = 0.0
Identities = 429/596 (71%), Positives = 497/596 (83%), Gaps = 2/596 (0%)
Frame = +2
Query: 464 YKDPKQPVAVRIKDLLGRMTLAEKIGQMTQIERENATADALAKYFIGSVLSGGGSVXXXX 643
Y+D + V VR++DLL RMTLAEK+GQMTQIER A+ AL Y+IGS+LSGGGSV
Sbjct: 34 YQDASKDVEVRVRDLLARMTLAEKVGQMTQIERIVASPQALRDYYIGSLLSGGGSVPRKQ 93
Query: 644 XXXXXXXXMVTEMQKGALSTRLGIPIIYGIDAVHGHNNVYKATIFPHNVGLGATRDPDLV 823
MV++ QK LSTRLGIP+IYGIDAVHGHNNVY ATIFPHNVGLGATRDP+LV
Sbjct: 94 ATAAEWVAMVSDFQKACLSTRLGIPMIYGIDAVHGHNNVYGATIFPHNVGLGATRDPNLV 153
Query: 824 KRIGEATALEVRATGIPYAFAPCIAVCRDPRWGRCYESYSEDPKVVQSLTSLISGLQGDA 1003
KRIG ATALEVRATGI YAFAPCIAVCRDPRWGRCYESYSED ++VQ++T LI GLQGD
Sbjct: 154 KRIGAATALEVRATGIQYAFAPCIAVCRDPRWGRCYESYSEDHRIVQAMTELIPGLQGDV 213
Query: 1004 PAD-SAGRPYVGGSKKVAACAKHYVGDGGTHNGINENNTIIDTHGLLSIHMPPYYNSIIR 1180
P + ++G P+ G KVAACAKH+VGDGGT NGINENNTIID GL+SIHMP Y +++ +
Sbjct: 214 PQNFTSGMPFAAGKDKVAACAKHFVGDGGTQNGINENNTIIDRQGLISIHMPAYLDALRK 273
Query: 1181 GVSTVMVSYSSWNGVKMHANHFLVTDFLKNKLKFRGFVISDWEGIDRITTPPHANYSYSI 1360
G STVM+SYSSWNG+KMHANH L+T FLK++L F+GF ISDWEGIDR+T+PP ANYSYS+
Sbjct: 274 GFSTVMISYSSWNGLKMHANHNLITGFLKDRLNFQGFTISDWEGIDRVTSPPGANYSYSV 333
Query: 1361 EAGVGAGIDMIMVPFRYTEFIDDLTTQVQNKVIPMSRIDDAVYRILRVKFTMGLFENPYP 1540
+A + AG+DMIMVP Y FI LT V + +IPMSRIDDAV RILRVKFTMGLFENP P
Sbjct: 334 QASILAGLDMIMVPNNYQNFITILTGHVNSGLIPMSRIDDAVTRILRVKFTMGLFENPMP 393
Query: 1541 DSSLAGELGKQEHRELAREAVRKSLVLLKNGKSSYAPLLPLPKKAGKILVAGSHANDLGN 1720
D SLA +LGKQEHR+LAREAVRKSLVLLKNGK APLLPLPKKA +ILVAGSHA++LG
Sbjct: 394 DPSLADQLGKQEHRDLAREAVRKSLVLLKNGKPGDAPLLPLPKKAARILVAGSHADNLGY 453
Query: 1721 QCGGWTITWQGSSGNTTAGTTILSGIEATVDPSTQVVYSESPDSG-VLADKYDYAIVVVG 1897
QCGGWTI WQG +G TT GTT+L ++A VDPST+VV++ESPD+ V + + YAIV VG
Sbjct: 454 QCGGWTIEWQGDTGRTTVGTTVLDAVKAAVDPSTEVVFAESPDAEFVRSGGFSYAIVAVG 513
Query: 1898 EPPYAETFGDNLNLTIPAPGPSVIQSVCGAAKCVVVLISGRPLVVEPYLGDMDALVAAWL 2077
E PY ET GD++NLTIP PGPS +Q+VC A +CV VLISGRP+V++P+LG MDA+VAAWL
Sbjct: 514 EHPYTETKGDSMNLTIPDPGPSTVQTVCAAVRCVTVLISGRPVVIQPFLGAMDAVVAAWL 573
Query: 2078 PGSEGQGVADVLFGDYGFTGKLPRTWFKSVDQLPMNVGDAHYDPLFPFGFGLTTKG 2245
PG+EGQGV DVLFGDYGFTGKLPRTWF+SVDQLPMN GDAHYDPLFP GFGLTT+G
Sbjct: 574 PGTEGQGVTDVLFGDYGFTGKLPRTWFRSVDQLPMNYGDAHYDPLFPLGFGLTTQG 629
>sptr|Q9XEI3|Q9XEI3 Beta-D-glucan exohydrolase isoenzyme ExoI.
Length = 630
Score = 873 bits (2255), Expect = 0.0
Identities = 439/616 (71%), Positives = 498/616 (80%), Gaps = 3/616 (0%)
Frame = +2
Query: 413 LVLMFCLLALGRAEYLKYKDPKQPVAVRIKDLLGRMTLAEKIGQMTQIERENATADALAK 592
L+L + +L A+Y+ YKD +PV R+ DLLGRMTLAEKIGQMTQIER AT D L
Sbjct: 13 LLLFWAVLGGTDADYVLYKDATKPVEDRVADLLGRMTLAEKIGQMTQIERLVATPDVLRD 72
Query: 593 YFIGSVLSGGGSVXXXXXXXXXXXXMVTEMQKGALSTRLGIPIIYGIDAVHGHNNVYKAT 772
FIGS+LSGGGSV MV QK +STRLGIP+IYGIDAVHG NNVY AT
Sbjct: 73 NFIGSLLSGGGSVPRKGATAKEWQDMVDGFQKACMSTRLGIPMIYGIDAVHGQNNVYGAT 132
Query: 773 IFPHNVGLGATRDPDLVKRIGEATALEVRATGIPYAFAPCIAVCRDPRWGRCYESYSEDP 952
IFPHNVGLGATRDP LVKRIGEATALEVRATGI YAFAPCIAVCRDPRWGRCYESYSED
Sbjct: 133 IFPHNVGLGATRDPYLVKRIGEATALEVRATGIQYAFAPCIAVCRDPRWGRCYESYSEDR 192
Query: 953 KVVQSLTSLISGLQGDAPAD-SAGRPYVGGSKKVAACAKHYVGDGGTHNGINENNTIIDT 1129
++VQS+T LI GLQGD P D ++G P+V G KVAACAKH+VGDGGT +GINENNTII+
Sbjct: 193 RIVQSMTELIPGLQGDVPKDFTSGMPFVAGKNKVAACAKHFVGDGGTVDGINENNTIINR 252
Query: 1130 HGLLSIHMPPYYNSIIRGVSTVMVSYSSWNGVKMHANHFLVTDFLKNKLKFRGFVISDWE 1309
GL++IHMP Y N++ +GVSTVM+SYSSWNGVKMHAN LVT +LK+ LKF+GFVISDWE
Sbjct: 253 EGLMNIHMPAYKNAMDKGVSTVMISYSSWNGVKMHANQDLVTGYLKDTLKFKGFVISDWE 312
Query: 1310 GIDRITTPPHANYSYSIEAGVGAGIDMIMVPFRYTEFIDDLTTQVQNKVIPMSRIDDAVY 1489
GIDRITTP ++YSYS++A + AG+DMIMVP Y +FI LT V VIPMSRIDDAV
Sbjct: 313 GIDRITTPAGSDYSYSVKASILAGLDMIMVPNNYQQFISILTGHVNGGVIPMSRIDDAVT 372
Query: 1490 RILRVKFTMGLFENPYPDSSLAGELGKQEHRELAREAVRKSLVLLKNGKSSY-APLLPLP 1666
RILRVKFTMGLFENPY D ++A +LGKQEHR+LAREA RKSLVLLKNGK+S APLLPLP
Sbjct: 373 RILRVKFTMGLFENPYADPAMAEQLGKQEHRDLAREAARKSLVLLKNGKTSTDAPLLPLP 432
Query: 1667 KKAGKILVAGSHANDLGNQCGGWTITWQGSSGNTTAGTTILSGIEATVDPSTQVVYSESP 1846
KKA KILVAGSHA++LG QCGGWTI WQG +G TT GTTIL ++A VDPST VV++E+P
Sbjct: 433 KKAPKILVAGSHADNLGYQCGGWTIEWQGDTGRTTVGTTILEAVKAAVDPSTVVVFAENP 492
Query: 1847 DSG-VLADKYDYAIVVVGEPPYAETFGDNLNLTIPAPGPSVIQSVCGAAKCVVVLISGRP 2023
D+ V + + YAIV VGE PY ET GDNLNLTIP PG S +Q+VCG +C VLISGRP
Sbjct: 493 DAEFVKSGGFSYAIVAVGEHPYTETKGDNLNLTIPEPGLSTVQAVCGGVRCATVLISGRP 552
Query: 2024 LVVEPYLGDMDALVAAWLPGSEGQGVADVLFGDYGFTGKLPRTWFKSVDQLPMNVGDAHY 2203
+VV+P L DALVAAWLPGSEGQGV D LFGD+GFTG+LPRTWFKSVDQLPMNVGDAHY
Sbjct: 553 VVVQPLLAASDALVAAWLPGSEGQGVTDALFGDFGFTGRLPRTWFKSVDQLPMNVGDAHY 612
Query: 2204 DPLFPFGFGLTTKGTK 2251
DPLF G+GLTT TK
Sbjct: 613 DPLFRLGYGLTTNATK 628
>sptrnew|AAQ97669|AAQ97669 Beta-glucanase.
Length = 633
Score = 870 bits (2248), Expect = 0.0
Identities = 427/627 (68%), Positives = 495/627 (78%), Gaps = 8/627 (1%)
Frame = +2
Query: 386 MASVHKATTLVLMFCLLALGRAEYLKYKDPKQPVAVRIKDLLGRMTLAEKIGQMTQIERE 565
M +V A +++ LL A+Y+KYKD KQP+ R++DLLGRMTL EKIGQM+QIER
Sbjct: 1 MTTVSSAAACLVLAVLLLPSAAQYVKYKDAKQPINERVQDLLGRMTLEEKIGQMSQIERA 60
Query: 566 NATADALAKYFIGSVLSGGGSVXXXXXXXXXXXXMVTEMQKGALSTRLGIPIIYGIDAVH 745
NATA+ + KYF+GSVLSGGGSV MVT MQK AL TRLGIPIIYGIDAVH
Sbjct: 61 NATAEVIEKYFVGSVLSGGGSVPAEKASASVWQKMVTRMQKAALKTRLGIPIIYGIDAVH 120
Query: 746 GHNNVYKATIFPHNVGLGATRDPDLVKRIGEATALEVRATGIPYAFAPCIAVCRDPRWGR 925
G+N+VY ATIFPHNVGLGATRDP LVKR+GEATA E RATGIPY FAPC+AVCRDPRWGR
Sbjct: 121 GNNDVYNATIFPHNVGLGATRDPRLVKRVGEATAHETRATGIPYTFAPCVAVCRDPRWGR 180
Query: 926 CYESYSEDPKVVQSLTS-LISGLQGDAPADSA-GRPYVGGSKKVAACAKHYVGDGGTHNG 1099
CYES+SED ++VQ +TS +++GLQGD PA G P+VGG+KKVA CAKH+VGDGGT G
Sbjct: 181 CYESFSEDTRLVQLMTSNMVAGLQGDVPAKHPKGVPFVGGAKKVAGCAKHFVGDGGTTRG 240
Query: 1100 INENNTIIDTHGLLSIHMPPYYNSIIRGVSTVMVSYSSWNGVKMHANHFLVTDFLKNKLK 1279
INENNT++ H L+ IHMPPY N++I G+S+VM+SYSSWNGVKMH N FL+TD LKNKL
Sbjct: 241 INENNTVLSFHDLMRIHMPPYDNAVINGISSVMISYSSWNGVKMHENKFLITDTLKNKLN 300
Query: 1280 FRGFVISDWEGIDRITTPPHANYSYSIEAGVGAGIDMIMVPFRYTEFIDDLTTQVQNKVI 1459
FRGFVI+DW+ +DRIT PPH +Y +SI+ + AGIDM+M+P+ Y EF+ DL QV+ I
Sbjct: 301 FRGFVITDWQAVDRITNPPHQHYYHSIKETIHAGIDMVMIPYDYPEFVADLAKQVKQGQI 360
Query: 1460 PMSRIDDAVYRILRVKFTMGLFENPYPDSSLAGELGKQEHRELAREAVRKSLVLLKNGKS 1639
+ RIDDAV RILRVKF MGLFE+P PD L ELG QEHR LAREAVRKSLVLLKN K
Sbjct: 361 KLERIDDAVSRILRVKFAMGLFEDPLPDPRLTKELGAQEHRALAREAVRKSLVLLKNSKK 420
Query: 1640 SYA-PLLPLPKKAGKILVAGSHANDLGNQCGGWTITWQGSSGN--TTAGTTILSGIEATV 1810
A P+LPLPK A KILVAGSHA+DLG+QCGGWTI WQG GN T GTTIL I+ V
Sbjct: 421 GQAKPMLPLPKTAKKILVAGSHAHDLGSQCGGWTIKWQGERGNNLTGVGTTILEAIKKAV 480
Query: 1811 DPSTQVVYSESPDSGVL---ADKYDYAIVVVGEPPYAETFGDNLNLTIPAPGPSVIQSVC 1981
D T V Y E PD L A+ Y+YA+V VGEPPYAET GDN NLTIP+PGP VI+ VC
Sbjct: 481 DKKTSVDYVERPDKDDLAKSAEGYEYAVVAVGEPPYAETAGDNKNLTIPSPGPEVIKDVC 540
Query: 1982 GAAKCVVVLISGRPLVVEPYLGDMDALVAAWLPGSEGQGVADVLFGDYGFTGKLPRTWFK 2161
G +CVV+++SGRPLV++PY+ MDALVAAWLPG+E QG+ DVLFGDYGFTGKLPRTWFK
Sbjct: 541 GLVRCVVLVVSGRPLVLQPYVDYMDALVAAWLPGTEAQGITDVLFGDYGFTGKLPRTWFK 600
Query: 2162 SVDQLPMNVGDAHYDPLFPFGFGLTTK 2242
SVDQLPMN GD YDPLFPFGFGLTTK
Sbjct: 601 SVDQLPMNYGDKRYDPLFPFGFGLTTK 627
>sptr|Q8RWM4|Q8RWM4 Beta-glucosidase-like protein.
Length = 626
Score = 850 bits (2195), Expect = 0.0
Identities = 422/614 (68%), Positives = 495/614 (80%), Gaps = 4/614 (0%)
Frame = +2
Query: 413 LVLMFCLLALGRAEYL--KYKDPKQPVAVRIKDLLGRMTLAEKIGQMTQIERENATADAL 586
L+L+ C +A + KYKDPK+P+ VRIK+L+ MTL EKIGQM Q+ER NAT + +
Sbjct: 13 LLLLCCTVAANKVPLANAKYKDPKEPLGVRIKNLMSHMTLEEKIGQMVQVERVNATTEVM 72
Query: 587 AKYFIGSVLSGGGSVXXXXXXXXXXXXMVTEMQKGALSTRLGIPIIYGIDAVHGHNNVYK 766
KYF+GSV SGGGSV MV E+QK ALSTRLGIPIIYGIDAVHGHN VY
Sbjct: 73 QKYFVGSVFSGGGSVPKPYIGPEAWVNMVNEVQKKALSTRLGIPIIYGIDAVHGHNTVYN 132
Query: 767 ATIFPHNVGLGATRDPDLVKRIGEATALEVRATGIPYAFAPCIAVCRDPRWGRCYESYSE 946
ATIFPHNVGLG TRDP LVKRIGEATALEVRATGI Y FAPCIAVCRDPRWGRCYESYSE
Sbjct: 133 ATIFPHNVGLGVTRDPGLVKRIGEATALEVRATGIQYVFAPCIAVCRDPRWGRCYESYSE 192
Query: 947 DPKVVQSLTSLISGLQGDAPADSAGRPYVGGSKKVAACAKHYVGDGGTHNGINENNTIID 1126
D K+VQ +T +I GLQGD P G P+V G KVAACAKH+VGDGGT G+N NNT+I+
Sbjct: 193 DHKIVQQMTEIIPGLQGDLPTGQKGVPFVAGKTKVAACAKHFVGDGGTLRGMNANNTVIN 252
Query: 1127 THGLLSIHMPPYYNSIIRGVSTVMVSYSSWNGVKMHANHFLVTDFLKNKLKFRGFVISDW 1306
++GLL IHMP Y++++ +GV+TVMVSYSS NG+KMHAN L+T FLKNKLKFRG VISD+
Sbjct: 253 SNGLLGIHMPAYHDAVNKGVATVMVSYSSINGLKMHANKKLITGFLKNKLKFRGIVISDY 312
Query: 1307 EGIDRITTPPHANYSYSIEAGVGAGIDMIMVPFRYTEFIDDLTTQVQNKVIPMSRIDDAV 1486
G+D+I TP ANYS+S+ A AG+DM M T+ ID+LT+QV+ K IPMSRIDDAV
Sbjct: 313 LGVDQINTPLGANYSHSVYAATTAGLDMFMGSSNLTKLIDELTSQVKRKFIPMSRIDDAV 372
Query: 1487 YRILRVKFTMGLFENPYPDSSLAGELGKQEHRELAREAVRKSLVLLKNGKSSYAPLLPLP 1666
RILRVKFTMGLFENP D SLA +LG +EHRELAREAVRKSLVLLKNG+++ PLLPLP
Sbjct: 373 KRILRVKFTMGLFENPIADHSLAKKLGSKEHRELAREAVRKSLVLLKNGENADKPLLPLP 432
Query: 1667 KKAGKILVAGSHANDLGNQCGGWTITWQGSSGNT-TAGTTILSGIEATVDPSTQVVYSES 1843
KKA KILVAG+HA++LG QCGGWTITWQG +GN T GTTIL+ ++ TVDP TQV+Y+++
Sbjct: 433 KKANKILVAGTHADNLGYQCGGWTITWQGLNGNNLTIGTTILAAVKKTVDPKTQVIYNQN 492
Query: 1844 PDSG-VLADKYDYAIVVVGEPPYAETFGDNLNLTIPAPGPSVIQSVCGAAKCVVVLISGR 2020
PD+ V A +DYAIV VGE PYAE FGD+ NLTI PGPS I +VC + KCVVV++SGR
Sbjct: 493 PDTNFVKAGDFDYAIVAVGEKPYAEGFGDSTNLTISEPGPSTIGNVCASVKCVVVVVSGR 552
Query: 2021 PLVVEPYLGDMDALVAAWLPGSEGQGVADVLFGDYGFTGKLPRTWFKSVDQLPMNVGDAH 2200
P+V++ + ++DALVAAWLPG+EGQGVADVLFGDYGFTGKL RTWFK+VDQLPMNVGD H
Sbjct: 553 PVVMQ--ISNIDALVAAWLPGTEGQGVADVLFGDYGFTGKLARTWFKTVDQLPMNVGDPH 610
Query: 2201 YDPLFPFGFGLTTK 2242
YDPL+PFGFGL TK
Sbjct: 611 YDPLYPFGFGLITK 624
>sptr|Q8GXR7|Q8GXR7 Hypothetical protein.
Length = 568
Score = 764 bits (1974), Expect = 0.0
Identities = 364/528 (68%), Positives = 439/528 (83%), Gaps = 3/528 (0%)
Frame = +2
Query: 668 MVTEMQKGALSTRLGIPIIYGIDAVHGHNNVYKATIFPHNVGLGATRDPDLVKRIGEATA 847
M+ E QKGAL +RLGIP+IYGIDAVHGHNNVY ATIFPHNVGLGATRDPDLVKRIG ATA
Sbjct: 1 MINEYQKGALVSRLGIPMIYGIDAVHGHNNVYNATIFPHNVGLGATRDPDLVKRIGAATA 60
Query: 848 LEVRATGIPYAFAPCIAVCRDPRWGRCYESYSEDPKVVQSLTSLISGLQGDAPAD-SAGR 1024
+EVRATGIPY FAPCIAVCRDPRWGRCYESYSED KVV+ +T +I GLQG+ P++ G
Sbjct: 61 VEVRATGIPYTFAPCIAVCRDPRWGRCYESYSEDHKVVEDMTDVILGLQGEPPSNYKHGV 120
Query: 1025 PYVGGSKKVAACAKHYVGDGGTHNGINENNTIIDTHGLLSIHMPPYYNSIIRGVSTVMVS 1204
P+VGG KVAACAKHYVGDGGT G+NENNT+ D HGLLS+HMP Y +++ +GVSTVMVS
Sbjct: 121 PFVGGRDKVAACAKHYVGDGGTTRGVNENNTVTDLHGLLSVHMPAYADAVYKGVSTVMVS 180
Query: 1205 YSSWNGVKMHANHFLVTDFLKNKLKFRGFVISDWEGIDRITTPPHANYSYSIEAGVGAGI 1384
YSSWNG KMHAN L+T +LK LKF+GFVISDW+G+D+I+TPPH +Y+ S+ A + AGI
Sbjct: 181 YSSWNGEKMHANTELITGYLKGTLKFKGFVISDWQGVDKISTPPHTHYTASVRAAIQAGI 240
Query: 1385 DMIMVPFRYTEFIDDLTTQVQNKVIPMSRIDDAVYRILRVKFTMGLFENPYPDSSLAGEL 1564
DM+MVPF +TEF++DLTT V+N IP++RIDDAV RIL VKFTMGLFENP D S + EL
Sbjct: 241 DMVMVPFNFTEFVNDLTTLVKNNSIPVTRIDDAVRRILLVKFTMGLFENPLADYSFSSEL 300
Query: 1565 GKQEHRELAREAVRKSLVLLKNGKSSYAPLLPLPKKAGKILVAGSHANDLGNQCGGWTIT 1744
G Q HR+LAREAVRKSLVLLKNG + P+LPLP+K KILVAG+HA++LG QCGGWTIT
Sbjct: 301 GSQAHRDLAREAVRKSLVLLKNGNKT-NPMLPLPRKTSKILVAGTHADNLGYQCGGWTIT 359
Query: 1745 WQGSSGN-TTAGTTILSGIEATVDPSTQVVYSESPDSG-VLADKYDYAIVVVGEPPYAET 1918
WQG SGN T GTT+LS +++ VD ST+VV+ E+PD+ + ++ + YAI+ VGEPPYAET
Sbjct: 360 WQGFSGNKNTRGTTLLSAVKSAVDQSTEVVFRENPDAEFIKSNNFAYAIIAVGEPPYAET 419
Query: 1919 FGDNLNLTIPAPGPSVIQSVCGAAKCVVVLISGRPLVVEPYLGDMDALVAAWLPGSEGQG 2098
GD+ LT+ PGP++I S C A KCVVV+ISGRPLV+EPY+ +DALVAAWLPG+EGQG
Sbjct: 420 AGDSDKLTMLDPGPAIISSTCQAVKCVVVVISGRPLVMEPYVASIDALVAAWLPGTEGQG 479
Query: 2099 VADVLFGDYGFTGKLPRTWFKSVDQLPMNVGDAHYDPLFPFGFGLTTK 2242
+ D LFGD+GF+GKLP TWF++ +QLPM+ GD HYDPLF +G GL T+
Sbjct: 480 ITDALFGDHGFSGKLPVTWFRNTEQLPMSYGDTHYDPLFAYGSGLETE 527
>sptr|Q9SD73|Q9SD73 Beta-D-glucan exohydrolase-like protein
(At3g47000).
Length = 608
Score = 716 bits (1847), Expect = 0.0
Identities = 354/597 (59%), Positives = 437/597 (73%), Gaps = 4/597 (0%)
Frame = +2
Query: 464 YKDPKQPVAVRIKDLLGRMTLAEKIGQMTQIERENATADALAKYFIGSVLSGGGSVXXXX 643
YK+ PV R+KDLL RMTL EKIGQMTQIER A+ A +FIGSVL+ GGSV
Sbjct: 10 YKNGDAPVEARVKDLLSRMTLPEKIGQMTQIERRVASPSAFTDFFIGSVLNAGGSVPFED 69
Query: 644 XXXXXXXXMVTEMQKGALSTRLGIPIIYGIDAVHGHNNVYKATIFPHNVGLGATRDPDLV 823
M+ Q+ AL++RLGIPIIYG DAVHG+NNVY AT+FPHN+GLGATRD DLV
Sbjct: 70 AKSSDWADMIDGFQRSALASRLGIPIIYGTDAVHGNNNVYGATVFPHNIGLGATRDADLV 129
Query: 824 KRIGEATALEVRATGIPYAFAPCIAVCRDPRWGRCYESYSEDPKVVQSLTSLISGLQGDA 1003
+RIG ATALEVRA+G+ +AF+PC+AV RDPRWGRCYESY EDP++V +TSL+SGLQG
Sbjct: 130 RRIGAATALEVRASGVHWAFSPCVAVLRDPRWGRCYESYGEDPELVCEMTSLVSGLQGVP 189
Query: 1004 PADSA-GRPYVGGSKKVAACAKHYVGDGGTHNGINENNTIIDTHGLLSIHMPPYYNSIIR 1180
P + G P+V G V AC KH+VGDGGT GINE NTI L IH+PPY + +
Sbjct: 190 PEEHPNGYPFVAGRNNVVACVKHFVGDGGTDKGINEGNTIASYEELEKIHIPPYLKCLAQ 249
Query: 1181 GVSTVMVSYSSWNGVKMHANHFLVTDFLKNKLKFRGFVISDWEGIDRITTPPHANYSYSI 1360
GVSTVM SYSSWNG ++HA+ FL+T+ LK KL F+GF++SDWEG+DR++ P +NY Y I
Sbjct: 250 GVSTVMASYSSWNGTRLHADRFLLTEILKEKLGFKGFLVSDWEGLDRLSEPQGSNYRYCI 309
Query: 1361 EAGVGAGIDMIMVPFRYTEFIDDLTTQVQNKVIPMSRIDDAVYRILRVKFTMGLFENPYP 1540
+ V AGIDM+MVPF+Y +FI D+T V++ IPM+RI+DAV RILRVKF GLF +P
Sbjct: 310 KTAVNAGIDMVMVPFKYEQFIQDMTDLVESGEIPMARINDAVERILRVKFVAGLFGHPLT 369
Query: 1541 DSSLAGELGKQEHRELAREAVRKSLVLLKNGKSSYAPLLPLPKKAGKILVAGSHANDLGN 1720
D SL +G +EHRELA+EAVRKSLVLLK+GK++ P LPL + A +ILV G+HA+DLG
Sbjct: 370 DRSLLPTVGCKEHRELAQEAVRKSLVLLKSGKNADKPFLPLDRNAKRILVTGTHADDLGY 429
Query: 1721 QCGGWTITWQGSSGNTTAGTTILSGIEATVDPSTQVVYSESPDSGVLA--DKYDYAIVVV 1894
QCGGWT TW G SG T GTT+L I+ V T+V+Y ++P LA + + YAIV V
Sbjct: 430 QCGGWTKTWFGLSGRITIGTTLLDAIKEAVGDETEVIYEKTPSKETLASSEGFSYAIVAV 489
Query: 1895 GEPPYAETFGDNLNLTIPAPGPSVIQSVCGAAKCVVVLISGRPLVVEP-YLGDMDALVAA 2071
GEPPYAET GDN L IP G ++ +V +V+LISGRP+V+EP L +ALVAA
Sbjct: 490 GEPPYAETMGDNSELRIPFNGTDIVTAVAEIIPTLVILISGRPVVLEPTVLEKTEALVAA 549
Query: 2072 WLPGSEGQGVADVLFGDYGFTGKLPRTWFKSVDQLPMNVGDAHYDPLFPFGFGLTTK 2242
WLPG+EGQGVADV+FGDY F GKLP +WFK V+ LP++ YDPLFPFGFGL +K
Sbjct: 550 WLPGTEGQGVADVVFGDYDFKGKLPVSWFKHVEHLPLDAHANSYDPLFPFGFGLNSK 606
>sptr|Q9LZJ4|Q9LZJ4 Beta-D-glucan exohydrolase-like protein.
Length = 650
Score = 703 bits (1814), Expect = 0.0
Identities = 365/626 (58%), Positives = 441/626 (70%), Gaps = 31/626 (4%)
Frame = +2
Query: 455 YLKYKDPKQPVAVRIKDLLGRMTLAEKIGQMTQIEREN----------ATADALAKYFIG 604
Y+KYKDPK V R++DLL RMTL EK+GQM QI+R N + KY IG
Sbjct: 35 YIKYKDPKVAVEERVEDLLIRMTLPEKLGQMCQIDRFNFSQVTGGVATVVPEIFTKYMIG 94
Query: 605 SVLSGGGSVXXXXXXXXXXXXMVTEMQKGALSTRLGIPIIYGIDAVHGHNNVYKATIFPH 784
SVLS + M+K +LSTRLGIP++Y +DAVHGHN ATIFPH
Sbjct: 95 SVLS---NPYDTGKDIAKRIFQTNAMKKLSLSTRLGIPLLYAVDAVHGHNTFIDATIFPH 151
Query: 785 NVGLGATRDPDLVKRIGEATALEVRATGIPYAFAPCIAVCRDPRWGRCYESYSEDPKVVQ 964
NVGLGATRDP LVK+IG TA EVRATG+ AFAPC+AVCRDPRWGRCYESYSEDP VV
Sbjct: 152 NVGLGATRDPQLVKKIGAITAQEVRATGVAQAFAPCVAVCRDPRWGRCYESYSEDPAVVN 211
Query: 965 SLT-SLISGLQGDAPADSAGRPYVGGSK-KVAACAKHYVGDGGTHNGINENNTIIDTHGL 1138
+T S+I GLQG+AP Y+ K VA CAKH+VGDGGT NGINENNT+ D L
Sbjct: 212 MMTESIIDGLQGNAP-------YLADPKINVAGCAKHFVGDGGTINGINENNTVADNATL 264
Query: 1139 LSIHMPPYYNSIIRGVSTVMVSYSSWNGVKMHANHFLVTDFLKNKLKFRGFVISDWEGID 1318
IHMPP+ ++ +G++++M SYSS NGVKMHAN ++TD+LKN LKF+GFVISDW GID
Sbjct: 265 FGIHMPPFEIAVKKGIASIMASYSSLNGVKMHANRAMITDYLKNTLKFQGFVISDWLGID 324
Query: 1319 RITTPPHANYSYSIEAGVGAGIDMIMVPFRYTEFIDDLTTQVQNKVIPMSRIDDAVYRIL 1498
+IT +NY+YSIEA + AGIDM+MVP+ Y E+++ LT V IPMSRIDDAV RIL
Sbjct: 325 KITPIEKSNYTYSIEASINAGIDMVMVPWAYPEYLEKLTNLVNGGYIPMSRIDDAVRRIL 384
Query: 1499 RVKFTMGLFENPYPDSSL-AGELGKQEHRELAREAVRKSLVLLKNGKSSYAPLLPLPKKA 1675
RVKF++GLFEN D L E G + HRE+ REAVRKS+VLLKNGK+ ++PLPKK
Sbjct: 385 RVKFSIGLFENSLADEKLPTTEFGSEAHREVGREAVRKSMVLLKNGKTDADKIVPLPKKV 444
Query: 1676 GKILVAGSHANDLGNQCGGWTITWQGSSG-------NT--------TAGTTILSGIEATV 1810
KI+VAG HAND+G QCGG+++TWQG +G NT GTTIL I+ V
Sbjct: 445 KKIVVAGRHANDMGWQCGGFSLTWQGFNGTGEDMPTNTKHGLPTGKIKGTTILEAIQKAV 504
Query: 1811 DPSTQVVYSESP--DSGVLADKYDYAIVVVGEPPYAETFGDNLNLTIPAPGPSVIQSVCG 1984
DP+T+VVY E P D+ L Y IVVVGE PYAETFGD+ L I PGP + CG
Sbjct: 505 DPTTEVVYVEEPNQDTAKLHADAAYTIVVVGETPYAETFGDSPTLGITKPGPDTLSHTCG 564
Query: 1985 AA-KCVVVLISGRPLVVEPYLGDMDALVAAWLPGSEGQGVADVLFGDYGFTGKLPRTWFK 2161
+ KC+V+L++GRPLV+EPY+ +DAL AWLPG+EGQGVADVLFGD+ FTG LPRTW K
Sbjct: 565 SGMKCLVILVTGRPLVIEPYIDMLDALAVAWLPGTEGQGVADVLFGDHPFTGTLPRTWMK 624
Query: 2162 SVDQLPMNVGDAHYDPLFPFGFGLTT 2239
V QLPMNVGD +YDPL+PFG+G+ T
Sbjct: 625 HVTQLPMNVGDKNYDPLYPFGYGIKT 650
>sptr|Q9SD72|Q9SD72 Beta-D-glucan exohydrolase-like protein.
Length = 609
Score = 697 bits (1800), Expect = 0.0
Identities = 345/596 (57%), Positives = 432/596 (72%), Gaps = 4/596 (0%)
Frame = +2
Query: 464 YKDPKQPVAVRIKDLLGRMTLAEKIGQMTQIERENATADALAKYFIGSVLSGGGSVXXXX 643
YK+ PV R+KDLL RMTL EKIGQMTQIER A+ + FIGSV SG GS
Sbjct: 11 YKNRDAPVEARVKDLLSRMTLPEKIGQMTQIERSVASPQVITNSFIGSVQSGAGSWPLED 70
Query: 644 XXXXXXXXMVTEMQKGALSTRLGIPIIYGIDAVHGHNNVYKATIFPHNVGLGATRDPDLV 823
M+ Q+ AL++RLGIPIIYG DAVHG+NNVY AT+FPHN+GLGATRD DLV
Sbjct: 71 AKSSDWADMIDGFQRSALASRLGIPIIYGTDAVHGNNNVYGATVFPHNIGLGATRDADLV 130
Query: 824 KRIGEATALEVRATGIPYAFAPCIAVCRDPRWGRCYESYSEDPKVVQSLTSLISGLQGDA 1003
KRIG ATALE+RA+G+ + FAPC+AV DPRWGRCYESYSE K+V ++ LISGLQG+
Sbjct: 131 KRIGAATALEIRASGVHWTFAPCVAVLGDPRWGRCYESYSEAAKIVCEMSLLISGLQGEP 190
Query: 1004 PADSA-GRPYVGGSKKVAACAKHYVGDGGTHNGINENNTIIDTHGLLSIHMPPYYNSIIR 1180
P + G P++ G V ACAKH+VGDGGT G++E NTI L IH+ PY N I +
Sbjct: 191 PEEHPYGYPFLAGRNNVIACAKHFVGDGGTEKGLSEGNTITSYEDLEKIHVAPYLNCIAQ 250
Query: 1181 GVSTVMVSYSSWNGVKMHANHFLVTDFLKNKLKFRGFVISDWEGIDRITTPPHANYSYSI 1360
GVSTVM S+SSWNG ++H+++FL+T+ LK KL F+GF++SDW+G++ I+ P +NY +
Sbjct: 251 GVSTVMASFSSWNGSRLHSDYFLLTEVLKQKLGFKGFLVSDWDGLETISEPEGSNYRNCV 310
Query: 1361 EAGVGAGIDMIMVPFRYTEFIDDLTTQVQNKVIPMSRIDDAVYRILRVKFTMGLFENPYP 1540
+ G+ AGIDM+MVPF+Y +FI D+T V++ IPM+R++DAV RILRVKF GLFE+P
Sbjct: 311 KLGINAGIDMVMVPFKYEQFIQDMTDLVESGEIPMARVNDAVERILRVKFVAGLFEHPLA 370
Query: 1541 DSSLAGELGKQEHRELAREAVRKSLVLLKNGKSSYAPLLPLPKKAGKILVAGSHANDLGN 1720
D SL G +G +EHRE+AREAVRKSLVLLKNGK++ P LPL + A +ILV G HANDLGN
Sbjct: 371 DRSLLGTVGCKEHREVAREAVRKSLVLLKNGKNADTPFLPLDRNAKRILVVGMHANDLGN 430
Query: 1721 QCGGWTITWQGSSGNTTAGTTILSGIEATVDPSTQVVYSESPDSGVLA--DKYDYAIVVV 1894
QCGGWT G SG T GTT+L I+A V T+V++ ++P LA D + YAIV V
Sbjct: 431 QCGGWTKIKSGQSGRITIGTTLLDSIKAAVGDKTEVIFEKTPTKETLASSDGFSYAIVAV 490
Query: 1895 GEPPYAETFGDNLNLTIPAPGPSVIQSVCGAAKCVVVLISGRPLVVEP-YLGDMDALVAA 2071
GEPPYAE GDN LTIP G ++I +V +V+L SGRP+V+EP L +ALVAA
Sbjct: 491 GEPPYAEMKGDNSELTIPFNGNNIITAVAEKIPTLVILFSGRPMVLEPTVLEKTEALVAA 550
Query: 2072 WLPGSEGQGVADVLFGDYGFTGKLPRTWFKSVDQLPMNVGDAHYDPLFPFGFGLTT 2239
W PG+EGQG++DV+FGDY F GKLP +WFK VDQLP+N YDPLFP GFGLT+
Sbjct: 551 WFPGTEGQGMSDVIFGDYDFKGKLPVSWFKRVDQLPLNAEANSYDPLFPLGFGLTS 606
>sptr|Q9SD69|Q9SD69 Beta-D-glucan exohydrolase-like protein.
Length = 636
Score = 688 bits (1775), Expect = 0.0
Identities = 351/626 (56%), Positives = 428/626 (68%), Gaps = 29/626 (4%)
Frame = +2
Query: 443 GRAEYLKYKDPKQPVAVRIKDLLGRMTLAEKIGQMTQIERENATADALAKYFIGSVLSGG 622
G E YK+ PV R+KDLL RMTL EKIGQMTQIER T + FIGSVL+GG
Sbjct: 3 GSNETCVYKNKDAPVEARVKDLLSRMTLPEKIGQMTQIERVVTTPPVITDNFIGSVLNGG 62
Query: 623 GSVXXXXXXXXXXXXMVTEMQKGALSTRLGIPIIYGIDAVHGHNNVYKATIFPHNVGLGA 802
GS M+ Q AL++RLGIPIIYGIDAVHG+NNVY ATIFPHN+GLGA
Sbjct: 63 GSWPFEDAKTSDWADMIDGYQNAALASRLGIPIIYGIDAVHGNNNVYGATIFPHNIGLGA 122
Query: 803 T-------------------------RDPDLVKRIGEATALEVRATGIPYAFAPCIAVCR 907
T RD DL++R+G ATALEVRA G +AFAPC+A R
Sbjct: 123 TSLVMLLHIDLEPKSLGRNKVVVKCDRDADLIRRVGAATALEVRACGAHWAFAPCVAALR 182
Query: 908 DPRWGRCYESYSEDPKVVQSLTSLISGLQGDAPADSA-GRPYVGGSKKVAACAKHYVGDG 1084
DPRWGR YESYSEDP ++ L+SL+SGLQG+ P + G P++ G V ACAKH+VGDG
Sbjct: 183 DPRWGRSYESYSEDPDIICELSSLVSGLQGEPPKEHPNGYPFLAGRNNVVACAKHFVGDG 242
Query: 1085 GTHNGINENNTIIDTHGLLSIHMPPYYNSIIRGVSTVMVSYSSWNGVKMHANHFLVTDFL 1264
GT GINE NTI+ L IH+ PY N + +GVSTVM SYSSWNG K+H+++FL+T+ L
Sbjct: 243 GTDKGINEGNTIVSYEELEKIHLAPYLNCLAQGVSTVMASYSSWNGSKLHSDYFLLTELL 302
Query: 1265 KNKLKFRGFVISDWEGIDRITTPPHANYSYSIEAGVGAGIDMIMVPFRYTEFIDDLTTQV 1444
K KL F+GFVISDWE ++R++ P +NY ++ V AG+DM+MVPF+Y +FI DLT V
Sbjct: 303 KQKLGFKGFVISDWEALERLSEPFGSNYRNCVKISVNAGVDMVMVPFKYEQFIKDLTDLV 362
Query: 1445 QNKVIPMSRIDDAVYRILRVKFTMGLFENPYPDSSLAGELGKQEHRELAREAVRKSLVLL 1624
++ + MSRIDDAV RILRVKF GLFE+P D SL G +G +EHRELARE+VRKSLVLL
Sbjct: 363 ESGEVTMSRIDDAVERILRVKFVAGLFEHPLTDRSLLGTVGCKEHRELARESVRKSLVLL 422
Query: 1625 KNGKSSYAPLLPLPKKAGKILVAGSHANDLGNQCGGWTITWQGSSGNTTAGTTILSGIEA 1804
KNG +S P LPL + +ILV G+HA+DLG QCGGWT W G SG T GTT+L I+
Sbjct: 423 KNGTNSEKPFLPLDRNVKRILVTGTHADDLGYQCGGWTKAWFGLSGRITIGTTLLDAIKE 482
Query: 1805 TVDPSTQVVYSESPDSGVLA--DKYDYAIVVVGEPPYAETFGDNLNLTIPAPGPSVIQSV 1978
V T+V+Y ++P LA ++ YAIV VGE PYAET GDN LTIP G ++ ++
Sbjct: 483 AVGDKTEVIYEKTPSEETLASLQRFSYAIVAVGETPYAETLGDNSELTIPLNGNDIVTAL 542
Query: 1979 CGAAKCVVVLISGRPLVVEP-YLGDMDALVAAWLPGSEGQGVADVLFGDYGFTGKLPRTW 2155
+VVL SGRPLV+EP L +ALVAAWLPG+EGQG+ DV+FGDY F GKLP +W
Sbjct: 543 AEKIPTLVVLFSGRPLVLEPLVLEKAEALVAAWLPGTEGQGMTDVIFGDYDFEGKLPVSW 602
Query: 2156 FKSVDQLPMNVGDAHYDPLFPFGFGL 2233
FK VDQLP+ YDPLFP GFGL
Sbjct: 603 FKRVDQLPLTADANSYDPLFPLGFGL 628
>sptr|Q9SD68|Q9SD68 Beta-D-glucan exohydrolase-like protein.
Length = 612
Score = 677 bits (1746), Expect = 0.0
Identities = 343/594 (57%), Positives = 423/594 (71%), Gaps = 4/594 (0%)
Frame = +2
Query: 464 YKDPKQPVAVRIKDLLGRMTLAEKIGQMTQIERENATADALAKYFIGSVLSGGGSVXXXX 643
YK+ + PV R+KDLL RMTLAEKIGQMT IER A+ + + IGSVL+ G
Sbjct: 10 YKNREAPVEARVKDLLSRMTLAEKIGQMTLIERSVASEAVIRDFSIGSVLNRAGGWPFED 69
Query: 644 XXXXXXXXMVTEMQKGALSTRLGIPIIYGIDAVHGHNNVYKATIFPHNVGLGATRDPDLV 823
M+ Q+ AL +RLGIPIIYGIDAVHG+N+VY ATIFPHN+GLGATRD DLV
Sbjct: 70 AKSSNWADMIDGFQRSALESRLGIPIIYGIDAVHGNNDVYGATIFPHNIGLGATRDADLV 129
Query: 824 KRIGEATALEVRATGIPYAFAPCIAVCRDPRWGRCYESYSEDPKVVQSLTSLISGLQGDA 1003
KRIG ATALEVRA G +AFAPC+AV +DPRWGRCYESY E ++V +TSL+SGLQG+
Sbjct: 130 KRIGAATALEVRACGAHWAFAPCVAVVKDPRWGRCYESYGEVAQIVSEMTSLVSGLQGEP 189
Query: 1004 PADSA-GRPYVGGSKKVAACAKHYVGDGGTHNGINENNTIIDTHGLLSIHMPPYYNSIIR 1180
D G P++ G K V ACAKH+VGDGGT+ INE NTI+ L H+ PY I +
Sbjct: 190 SKDHTNGYPFLAGRKNVVACAKHFVGDGGTNKAINEGNTILRYEDLERKHIAPYKKCISQ 249
Query: 1181 GVSTVMVSYSSWNGVKMHANHFLVTDFLKNKLKFRGFVISDWEGIDRITTPPHANYSYSI 1360
GVSTVM SYSSWNG K+H+++FL+T+ LK KL F+G+V+SDWEG+DR++ PP +NY +
Sbjct: 250 GVSTVMASYSSWNGDKLHSHYFLLTEILKQKLGFKGYVVSDWEGLDRLSDPPGSNYRNCV 309
Query: 1361 EAGVGAGIDMIMVPFRYTEFIDDLTTQVQNKVIPMSRIDDAVYRILRVKFTMGLFENPYP 1540
+ G+ AGIDM+MVPF+Y +F +DL V++ + M+R++DAV RILRVKF GLFE P
Sbjct: 310 KIGINAGIDMVMVPFKYEQFRNDLIDLVESGEVSMARVNDAVERILRVKFVAGLFEFPLT 369
Query: 1541 DSSLAGELGKQEHRELAREAVRKSLVLLKNGKSSYAPLLPLPKKAGKILVAGSHANDLGN 1720
D SL +G +EHRELAREAVRKSLVLLKNG+ Y LPL A +ILV G+HA+DLG
Sbjct: 370 DRSLLPTVGCKEHRELAREAVRKSLVLLKNGR--YGEFLPLNCNAERILVVGTHADDLGY 427
Query: 1721 QCGGWTITWQGSSGNTTAGTTILSGIEATVDPSTQVVYSESPDSGVLAD--KYDYAIVVV 1894
QCGGWT T G SG T GTT+L I+A V T+V+Y +SP LA ++ YAIV V
Sbjct: 428 QCGGWTKTMYGQSGRITDGTTLLDAIKAAVGDETEVIYEKSPSEETLASGYRFSYAIVAV 487
Query: 1895 GEPPYAETFGDNLNLTIPAPGPSVIQSVCGAAKCVVVLISGRPLVVEP-YLGDMDALVAA 2071
GE PYAET GDN L IP G +I +V +V+L SGRP+ +EP L +ALVAA
Sbjct: 488 GESPYAETMGDNSELVIPFNGSEIITTVAEKIPTLVILFSGRPMFLEPQVLEKAEALVAA 547
Query: 2072 WLPGSEGQGVADVLFGDYGFTGKLPRTWFKSVDQLPMNVGDAHYDPLFPFGFGL 2233
WLPG+EGQG+ADV+FGDY F GKLP TWFK VDQLP+++ Y PLFP GFGL
Sbjct: 548 WLPGTEGQGIADVIFGDYDFRGKLPATWFKRVDQLPLDIESNGYLPLFPLGFGL 601
>sptr|Q82JP6|Q82JP6 Putative glycosyl hydrolase.
Length = 1011
Score = 551 bits (1420), Expect = e-155
Identities = 303/616 (49%), Positives = 388/616 (62%), Gaps = 19/616 (3%)
Frame = +2
Query: 449 AEYLKYKDPKQPVAVRIKDLLGRMTLAEKIGQMTQIERENATADA-LAKYFIGSVLSGGG 625
A L Y DPK PV R+ DL+ RM+L EK GQMTQ ER TA +A Y +GS+LSGGG
Sbjct: 327 AHGLPYLDPKLPVKKRVADLVSRMSLEEKAGQMTQAERGALTAQGDIAAYDLGSLLSGGG 386
Query: 626 SVXXXXXXXXXXXXMVTEMQKGALSTRLGIPIIYGIDAVHGHNNVYKATIFPHNVGLGAT 805
S M+ Q A +TR IP+IYG+DAVHGHNN+ ATI PHN+G+GA+
Sbjct: 387 STPTPNTPEAWAK-MIDAFQLRAQATRFQIPLIYGVDAVHGHNNLTGATIMPHNIGIGAS 445
Query: 806 RDPDLVKRIGEATALEVRATGIPYAFAPCIAVCRDPRWGRCYESYSEDPKVVQSLTSLIS 985
RDP + +R G TA EVRATGIP+ FAPC+ V RD RWGR YES+ EDP +V+S+ ++I
Sbjct: 446 RDPGIARRTGAVTAAEVRATGIPWDFAPCLCVTRDERWGRSYESFGEDPALVKSMETVIQ 505
Query: 986 GLQGDAPADSAGRPYVGGSKKVAACAKHYVGDGGTHNG--------INENNTIIDTHGLL 1141
GLQG + KV A AKH+VGDGGT G I++ T + L
Sbjct: 506 GLQGARDGKD-----LKDDDKVLATAKHFVGDGGTAYGSSTTGTYTIDQGVTKVTRQELE 560
Query: 1142 SIHMPPYYNSIIRGVSTVMVSYSSWN-------GVKMHANHFLVTDFLKNKLKFRGFVIS 1300
++ + PY ++ RG+ +VM SYSS + VKMHA ++ LK+++ F GFVIS
Sbjct: 561 AVQLAPYQTAVDRGIGSVMPSYSSLDILGDGQGAVKMHARADMINGVLKDRMGFDGFVIS 620
Query: 1301 DWEGIDRITTPPHANYSYSIEAGVGAGIDMIMVPFRYTEFIDDLTTQVQNKVIPMSRIDD 1480
DW+ ID+I +Y+ + + AG+DMIMVP+ Y +F L +V+ + R+DD
Sbjct: 621 DWKAIDQIP----GDYASDVRTSINAGLDMIMVPYEYKDFRTTLIDEVKAGRVSQKRVDD 676
Query: 1481 AVYRILRVKFTMGLFENPYPDSSLAGELGKQEHRELAREAVRKSLVLLKNGKSSYAPLLP 1660
AV RIL KF +GLFE PY D+S A +G HR +AREA +S VLLKN +LP
Sbjct: 677 AVSRILTQKFKLGLFEKPYADTSGASRIGSSAHRAVAREAAAESQVLLKNA----GGVLP 732
Query: 1661 LPKKAGKILVAGSHANDLGNQCGGWTITWQGSSGNTTAGTTILSGIEATVDPSTQVVYSE 1840
L KK+ K+ VAGS+A+DLGNQ GGWTITWQGSSG T GTTIL G+ V YS+
Sbjct: 733 L-KKSQKVYVAGSNADDLGNQTGGWTITWQGSSGKHTDGTTILDGMRKAAGSGGAVTYSK 791
Query: 1841 SPDSGVLADKYDYAIVVVGEPPYAETFGDNLN---LTIPAPGPSVIQSVCGAAKCVVVLI 2011
D+ YD +VVVGE PYAE GD N L + A + + VCGA KC V+++
Sbjct: 792 --DASAPTSGYDVGVVVVGETPYAEGVGDVGNGNDLELTAADKAAVDKVCGAMKCAVLIV 849
Query: 2012 SGRPLVVEPYLGDMDALVAAWLPGSEGQGVADVLFGDYGFTGKLPRTWFKSVDQLPMNVG 2191
SGRP ++ LGD+DALVA+WLPG+EG GVADVL+G FTG+LP TW KS QLP+NVG
Sbjct: 850 SGRPQLIGDRLGDIDALVASWLPGTEGDGVADVLYGKRAFTGQLPVTWPKSEAQLPINVG 909
Query: 2192 DAHYDPLFPFGFGLTT 2239
D YDP FP+G+GLTT
Sbjct: 910 DTTYDPQFPYGWGLTT 925
>sptr|Q97K46|Q97K46 Beta-glucosidase family protein.
Length = 665
Score = 473 bits (1217), Expect = e-132
Identities = 277/620 (44%), Positives = 363/620 (58%), Gaps = 29/620 (4%)
Frame = +2
Query: 464 YKDPKQPVAVRIKDLLGRMTLAEKIGQMTQIE----RENATADALAKYFIGSVLSGGGSV 631
Y +P R+ DLL RMTL EK+GQM Q E +NA + Y +GSVLSGG +
Sbjct: 41 YLNPNANTEARVSDLLKRMTLDEKVGQMLQGELGTSSDNAKPEDCINYTLGSVLSGGNAD 100
Query: 632 XXXXXXXXXXXXMVTEMQKGALSTRLGIPIIYGIDAVHGHNNVYKATIFPHNVGLGATRD 811
VT + + RL IPI+YG+DAVHG++N+ ATIFPHN+ LGA
Sbjct: 101 PTTGNDAQSWYDTVTSFVEASTQNRLHIPILYGVDAVHGNSNIIGATIFPHNIALGAIAT 160
Query: 812 PDL------VKRIGEATALEVRATGIPYAFAPCIAVCRDPRWGRCYESYSEDPKVVQSL- 970
+L V++IG ATA E+R T IP+ FAPC+A ++P WGR YE + ED + L
Sbjct: 161 GNLNEGKKIVRKIGSATAKEMRVTDIPWTFAPCLANPQNPTWGRTYEGFGEDINLASELG 220
Query: 971 TSLISGLQGDAPADSAGRPYVGGSKKVAACAKHYVGDGGTHNGINENNTIIDTHGLLSIH 1150
+S I GLQG+ D + K A KHY+G+G T NG N+ N T ++ +
Sbjct: 221 SSYIKGLQGNDIND------LKKPNKAVATIKHYLGEGYTENGTNQGNVTSMTKEQVAKN 274
Query: 1151 M-PPYYNSIIRGVSTVMVSYSSWNGVKMHANHFLVTDFLKNKLKFRGFVISDWEGIDRIT 1327
+ PY +++ G TVM SY+S GVKM A+ +L+TD LKNKLKF GFVISD+ +IT
Sbjct: 275 LIKPYEDAVRAGARTVMPSYNSIQGVKMTASKYLLTDILKNKLKFDGFVISDYNAAQQIT 334
Query: 1328 TPPHAN----YSYSIEAGVGAGIDMIMVPFRYTEFIDDLTTQVQNKV------IPMSRID 1477
+ N ++ + AG+DM+M P + I + V ++ IPMSRI+
Sbjct: 335 ADENGNSVSGLKNQVKVSINAGVDMLMEPNDWKSCIGYIKELVADEKAHPGTGIPMSRIN 394
Query: 1478 DAVYRILRVKFTMGLFE-----NPYPDSSLAGELGKQEHRELAREAVRKSLVLLKNGKSS 1642
DAV RILRVKF GLFE NP + + +LG +HR+LAREAV KSLVLLKN
Sbjct: 395 DAVSRILRVKFQSGLFEHPISNNPENNPKVMAQLGSNKHRKLAREAVSKSLVLLKNDAVG 454
Query: 1643 YAPLLPLPKKAGKILVAGSHANDLGNQCGGWTITWQGSSGNTTAGTTILSGIEATVDPST 1822
P+L KK KI VAG AND+GNQCGGWTI WQG SGNTT GTTIL GI+ ++ P
Sbjct: 455 GKPILSQLKKMKKIFVAGKSANDIGNQCGGWTIDWQGKSGNTTKGTTILQGIKDSISPKQ 514
Query: 1823 QVVYSESPDSGVLADKYDYAIVVVGEPPYAETFGDNLN-LTIPAPGPSVIQSV-CGAAKC 1996
V +SE G A D AI ++GE PYAET GDNLN L + + + ++
Sbjct: 515 NVTFSE---DGAGASGNDVAIAIIGETPYAETNGDNLNGLNLDSTDKKTLANLKASGVPT 571
Query: 1997 VVVLISGRPLVVEPYLGDMDALVAAWLPGSEGQGVADVLFGDYGFTGKLPRTWFKSVDQL 2176
+VVL+SGRP++V Y+ D LV AWLPG+EG GV+DVLFG+ FTG+LP W +
Sbjct: 572 IVVLVSGRPMIVTDYIKDWAGLVEAWLPGTEGNGVSDVLFGNKDFTGRLPEKWAFYTEAY 631
Query: 2177 PMNVGDAHYDPLFPFGFGLT 2236
P+ + Y LF G+GLT
Sbjct: 632 PITNSNKQY-MLFDSGYGLT 650
>sptr|Q9AA14|Q9AA14 1,4-beta-D-glucan glucohydrolase D.
Length = 823
Score = 452 bits (1162), Expect = e-125
Identities = 267/605 (44%), Positives = 356/605 (58%), Gaps = 25/605 (4%)
Frame = +2
Query: 497 IKDLLGRMTLAEKIGQMTQIERENATADALAKYFIGSVLSGGGSVXXXXXXXXXXXXMVT 676
+ +L ++TL EK+GQ+ Q + + + L Y +GS+L GG S V
Sbjct: 52 VDSVLAKLTLEEKVGQLIQADIGSIKPEDLKTYPLGSILGGGSSPPLNAPDRSPQKPWVD 111
Query: 677 EMQ--KGALSTRLG---IPIIYGIDAVHGHNNVYKATIFPHNVGLGATRDPDLVKRIGEA 841
+ + A + R+G +P+I+GIDAVHG+NNV AT+FPHN+GLGA RDP+L++RIG+A
Sbjct: 112 TAKAFREAAAQRVGGTHVPLIFGIDAVHGNNNVVGATLFPHNIGLGAARDPELIRRIGKA 171
Query: 842 TALEVRATGIPYAFAPCIAVCRDPRWGRCYESYSEDPKVVQSL-TSLISGLQGDAPADSA 1018
TALE A G +AF P +A RD RWGR YE YSEDP +++ +I GLQG +
Sbjct: 172 TALETSAAGFDWAFGPTLAAPRDDRWGRTYEGYSEDPAIIRQYGGEMILGLQGAVAPFTG 231
Query: 1019 GRPYVGGSKKVAACAKHYVGDGGTHNGINENNTIIDTHGLLSIHMPPYYNSIIRGVSTVM 1198
G+ V VAA AKH++GDGGT G+++ +T + L+ IH Y +I G TVM
Sbjct: 232 GKAGVIQQGLVAASAKHFLGDGGTDKGVDQGDTKVSEEELVRIHAQGYVPAINAGALTVM 291
Query: 1199 VSYSSWNGVKMHANHFLVTDFLKNKLKFRGFVISDWEGIDRITTPPHANYSYSIEAGVGA 1378
S++SWNG KMH N L+TD LK K+ F GF++ DW G ++ + + SI A
Sbjct: 292 ASFNSWNGQKMHGNKSLMTDVLKGKMGFDGFIVGDWNGHGQVAGCKPTDCAQSI----NA 347
Query: 1379 GIDMIMVPFRYTEFIDDLTTQVQNKVIPMSRIDDAVYRILRVKFTMGLFENPYPDSSLAG 1558
G+DM M P + D+ Q ++ VIPM+RIDDAV RILRVK MGLF+ P G
Sbjct: 348 GLDMFMAPDSWKGLYDNTLAQAKSGVIPMARIDDAVRRILRVKAKMGLFQAARPYEGRQG 407
Query: 1559 ELGKQEHRELAREAVRKSLVLLKNGKSSYAPLLPLPKKAGKILVAGSHANDLGNQCGGWT 1738
+G EHR +AREAVRKSLVLLKN +LP+ K + +LVAGS A+D+G Q GGWT
Sbjct: 408 VIGAPEHRAIAREAVRKSLVLLKND-----GVLPV-KASANVLVAGSGADDIGKQSGGWT 461
Query: 1739 ITWQGSSGNTTAG--------TTILSGIEATVDPSTQVVYSESPDSGVLADKYDYAIVVV 1894
++WQG +GNT A T + S +EA +T V G K D AIVV
Sbjct: 462 LSWQG-TGNTNADFPNADSIWTGVKSAVEAGGGRATLSV------DGKFDKKPDVAIVVF 514
Query: 1895 GEPPYAETFGDNLNLTIPAPGP----SVIQSV-CGAAKCVVVLISGRPLVVEPYLGDMDA 2059
GE PYAE GD + PG ++++S+ K V V ++GRPL V P + DA
Sbjct: 515 GENPYAEGVGDLKSTLEYQPGAKADLALLKSLKAQGVKVVSVFLTGRPLWVNPEINASDA 574
Query: 2060 LVAAWLPGSEGQGVADVLFGD------YGFTGKLPRTWFKSVDQLPMNVGDAHYDPLFPF 2221
VAAWLPGSEG GVADVL GD + F GKL +W K+ Q +N GD YDPLF +
Sbjct: 575 FVAAWLPGSEGAGVADVLIGDKAGKPRHDFAGKLSFSWPKTAGQFRLNKGDKGYDPLFAY 634
Query: 2222 GFGLT 2236
G+GL+
Sbjct: 635 GYGLS 639
>sptr|Q9A6N5|Q9A6N5 1,4-beta-D-glucan glucohydrolase D.
Length = 821
Score = 441 bits (1135), Expect = e-122
Identities = 269/599 (44%), Positives = 349/599 (58%), Gaps = 19/599 (3%)
Frame = +2
Query: 497 IKDLLGRMTLAEKIGQMTQIERENATADALAKYFIGSVLSGGGSVXXXXXXXXXXXXM-- 670
I L+ RMT+ EK+ Q Q + + T + L KY +GSVL GG S +
Sbjct: 57 IAQLMSRMTVEEKVAQTIQADGASITPEELKKYRLGSVLVGGNSAPDGNDRASPQRWIEW 116
Query: 671 VTEMQKGALSTR---LGIPIIYGIDAVHGHNNVYKATIFPHNVGLGATRDPDLVKRIGEA 841
+ + AL R IPII+G+DAVHGHNNV ATIFPHNVGLGA +PDL++RIGE
Sbjct: 117 IRAFRAAALDKRGDRQEIPIIFGVDAVHGHNNVVGATIFPHNVGLGAAHEPDLIRRIGEV 176
Query: 842 TALEVRATGIPYAFAPCIAVCRDPRWGRCYESYSEDPKVVQSLTS-LISGLQGDAPADSA 1018
TA E+ ATG + F P +AV RD RWGR YE Y E+P++V++ + + GLQG A A
Sbjct: 177 TAKEMAATGADWTFGPTVAVPRDSRWGRAYEGYGENPEIVKAYSGPMTLGLQG---ALEA 233
Query: 1019 GRPYVGGSKKVAACAKHYVGDGGTHNGINENNTIIDTHGLLSIHMPPYYNSIIRGVSTVM 1198
G+P G +VA AKH++ DGGT NG ++ + I L+ +H Y +I G+ +VM
Sbjct: 234 GKPLAAG--RVAGSAKHFLADGGTENGRDQGDAKISEADLVRLHNAGYPPAIEAGILSVM 291
Query: 1199 VSYSSWNGVKMHANHFLVTDFLKNKLKFRGFVISDWEGIDRITTPPHANYSYSIEAGVGA 1378
VS+SSWNGVK N L+TD LK ++ F GFV+ DW ++ + S A
Sbjct: 292 VSFSSWNGVKHTGNKSLLTDVLKERMGFEGFVVGDWNAHGQV----EGCSNTSCAQAYNA 347
Query: 1379 GIDMIMVPFRYTEFIDDLTTQVQNKVIPMSRIDDAVYRILRVKFTMGLFENPYPDSSLAG 1558
G+DM+M P + D+ QV+ IPM+RIDDAV RILRVK GLFE+ P
Sbjct: 348 GMDMMMAPDSWKGLYDNTLAQVKAGQIPMARIDDAVRRILRVKVKAGLFEDKRPLEGKLE 407
Query: 1559 ELGKQEHRELAREAVRKSLVLLKNGKSSYAPLLPLPKKAGKILVAGSHANDLGNQCGGWT 1738
LG EHR +AREAVRKSLVLLKN +LPL K + ++LVAG A+D+G GGWT
Sbjct: 408 LLGAPEHRAVAREAVRKSLVLLKN-----EGVLPL-KSSARVLVAGDGADDIGKASGGWT 461
Query: 1739 ITWQGSSGNTT---AGTTILSGIEATVDPSTQVVYSESPDSGVLADKYDYAIVVVGEPPY 1909
+TWQG+ + G +I +G+ V +E SG K D AIVV GE PY
Sbjct: 462 LTWQGTGNKNSDFPHGQSIYAGVAEAVKAGGG--SAELSVSGDFKQKPDVAIVVFGENPY 519
Query: 1910 AETFGDNLNLTIPAPGP---SVIQSVCGAAKCVV-VLISGRPLVVEPYLGDMDALVAAWL 2077
AE GD ++ A ++++ + A VV V +SGRPL P L DA VAAWL
Sbjct: 520 AEFQGDITSIEYQAGDKRDLALLKKLKAAGIPVVSVFLSGRPLWTNPELNASDAFVAAWL 579
Query: 2078 PGSEGQGVADVLFGD------YGFTGKLPRTWFKSVDQLPMNVGDAHYDPLFPFGFGLT 2236
PGSEG GVADVL GD + F GKL +W K DQ P+NVGD YDPLF +G+GL+
Sbjct: 580 PGSEGGGVADVLVGDKAGKPRHDFQGKLSFSWPKRADQEPINVGDPGYDPLFAYGYGLS 638
>sptr|Q47912|Q47912 1,4-B-D-glucan glucohydrolase.
Length = 869
Score = 438 bits (1127), Expect = e-121
Identities = 269/617 (43%), Positives = 364/617 (58%), Gaps = 27/617 (4%)
Frame = +2
Query: 467 KDPKQPVAVRIKDLLGRMTLAEKIGQMTQIERENATADALAKYFIGSVLSGGGSVXXXXX 646
KDP V R+ DLL RMTL EKIGQ+ Q E + T + + +Y +GSVL+GGGS
Sbjct: 64 KDPA--VEARVDDLLARMTLEEKIGQLVQPEIRHVTPEDIKQYHVGSVLNGGGSTPGANK 121
Query: 647 XXX-------XXXXMVTEMQKGALSTRLGIPIIYGIDAVHGHNNVYKATIFPHNVGLGAT 805
+ K R+GIP+I+G DAVHG NV AT+FPHN+GLGAT
Sbjct: 122 YASLEDWCKLADSFYYASVDKS--DGRIGIPVIWGTDAVHGLGNVIGATLFPHNIGLGAT 179
Query: 806 RDPDLVKRIGEATALEVRATGIPYAFAPCIAVCRDPRWGRCYESYSEDPKVVQSLT-SLI 982
+P+L+K+IG ATA E+ ATG+ + F+P +AV RD RWGR YES+SEDP++V + ++
Sbjct: 180 NNPELLKQIGWATAREIAATGLDWYFSPTVAVARDDRWGRTYESWSEDPQIVHAFAGKMV 239
Query: 983 SGLQGDAPADSAGRPYVGGSKKVAACAKHYVGDGGTHNGINENNTIIDTHGLLSIHMPPY 1162
GLQG + G + + V A AKH++GDGGT NG++ T D L IH Y
Sbjct: 240 EGLQG-----TGGSDRLFTHEHVIATAKHFIGDGGTLNGVDRGETQGDEKVLRDIHGAGY 294
Query: 1163 YNSIIRGVSTVMVSYSSWNGVKMHANHFLVTDFLKNKLKFRGFVISDWEGIDRITTPPHA 1342
+++I GV VM S++SW G +MH + +L+TD LK+ F G V+ DW G I
Sbjct: 295 FSAIESGVQVVMASFTSWEGTRMHGHKYLLTDVLKDLSGFDGLVVGDWSGHSFIPGCTAL 354
Query: 1343 NYSYSIEAGVGAGIDMIMVPF-RYTEFIDDLTTQVQNKVIPMSRIDDAVYRILRVKFTMG 1519
N S+ AG+D+ MVP + E +L Q + +PM+R+DDAV ILRVK G
Sbjct: 355 NCPQSLM----AGLDIYMVPEPDWEELYKNLLAQAKTGELPMARVDDAVRAILRVKIRAG 410
Query: 1520 LFENPYPDS-SLAGE---LGKQEHRELAREAVRKSLVLLKNGKSSYAPLLPLPKKAGKIL 1687
LFE P + LAG+ LG EHRE+AR+AVR+SLVLLKN + LLPL ++ +L
Sbjct: 411 LFEKGAPSTRPLAGKKDVLGAPEHREVARQAVRESLVLLKNKNN----LLPLARQQ-TVL 465
Query: 1688 VAGSHANDLGNQCGGWTITWQGSSGNTTA----GTTILSGIEATVDPSTQVVYSESPDSG 1855
V G A++ G Q GGW+++WQG +GNT A T+I +GI A V+ + D G
Sbjct: 466 VTGDGADNSGKQSGGWSVSWQG-TGNTNADFPGATSIYAGINAVVEQAGGKTLLS--DDG 522
Query: 1856 VLADKYDYAIVVVGEPPYAETFGDNLNLTIPAPGPS---VIQSVCGAAKCVVVL-ISGRP 2023
++K D AIVV GE PYAE GD N+ S +++ + VV L ISGRP
Sbjct: 523 SFSEKPDVAIVVFGEDPYAEMQGDVGNMAYKPRDTSDWELLKKLRSQGIPVVSLFISGRP 582
Query: 2024 LVVEPYLGDMDALVAAWLPGSEGQGVADVLFG------DYGFTGKLPRTWFKSVDQLPMN 2185
L V + DA VA WLPG+EGQG+ADV+F +Y G+L +W K +Q P+N
Sbjct: 583 LWVNREINASDAFVAVWLPGTEGQGIADVIFRNAQGEINYDVKGRLSFSWPKRPEQTPLN 642
Query: 2186 VGDAHYDPLFPFGFGLT 2236
GDA+YDPLFP+G+GL+
Sbjct: 643 RGDANYDPLFPYGYGLS 659
>sptr|Q9F0N9|Q9F0N9 Cellobiase CelA precursor.
Length = 685
Score = 426 bits (1095), Expect = e-118
Identities = 262/613 (42%), Positives = 353/613 (57%), Gaps = 27/613 (4%)
Frame = +2
Query: 485 VAVRIKDLLGRMTLAEKIGQMTQIERENATADALAKYFIGSVLSGGGSVXXXXXXX--XX 658
V R+ LL ++++ EK+GQ+ Q + T + L KY +GS+L+GG S
Sbjct: 55 VEKRVDALLKQLSVEEKVGQVIQGDIGTITPEDLRKYPLGSILAGGNSGPNGDDRAPPKE 114
Query: 659 XXXMVTEMQKGALSTRLG---IPIIYGIDAVHGHNNVYKATIFPHNVGLGATRDPDLVKR 829
+ + +L R G IP+++GIDAVHGH N+ ATIFPHN+ LGAT DP+L++R
Sbjct: 115 WLDLADAFYRVSLEKRPGHTPIPVLFGIDAVHGHGNIGSATIFPHNIALGATHDPELLRR 174
Query: 830 IGEATALEVRATGIPYAFAPCIAVCRDPRWGRCYESYSEDPKVVQSLT-SLISGLQGDAP 1006
IGE TA+E+ ATGI + FAP ++V RD RWGR YE +SEDP++V + + +++ G+QG
Sbjct: 175 IGEVTAVEMAATGIDWTFAPALSVVRDDRWGRTYEGFSEDPEIVAAYSAAIVEGVQGKFG 234
Query: 1007 ADSAGRPYVGGSKKVAACAKHYVGDGGTHNGINENNTIIDTHGLLSIHMPPYYNSIIRGV 1186
+ P ++ A AKH++ DGGT G ++ + I L+ IH Y +I GV
Sbjct: 235 SKDFMAP-----GRIVASAKHFLADGGTDQGRDQGDARISEDELIRIHNAGYPPAIDAGV 289
Query: 1187 STVMVSYSSWNGVKMHANHFLVTDFLKNKLKFRGFVISDWEGIDRITTPPHANYSYSIEA 1366
TVM S+SSW G+K H + L+TD LK ++ F GF++ DW D++ N S+
Sbjct: 290 LTVMASFSSWQGIKHHGHKQLLTDVLKGQMGFNGFIVGDWNAHDQVPGCTKFNCPTSLI- 348
Query: 1367 GVGAGIDMIMVPFRYTEFIDDLTTQVQNKVIPMSRIDDAVYRILRVKFTMGLFENPYPDS 1546
AG+DM M + + ++ QV++ IPM+R+DDAV RILRVK GLFE P P
Sbjct: 349 ---AGLDMYMAADSWKQLYENTLAQVKDGTIPMARLDDAVRRILRVKVLAGLFEKPAPKD 405
Query: 1547 --SLAG--ELGKQEHRELAREAVRKSLVLLKNGKSSYAPLLPLPKKAGKILVAGSHANDL 1714
L G LG EHR + REAVRKSLVLLKN K + LPL KA ++LVAG A+++
Sbjct: 406 RPGLPGLETLGSPEHRAVGREAVRKSLVLLKNDKGT----LPLSPKA-RVLVAGDGADNI 460
Query: 1715 GNQCGGWTITWQGSSGNT---TAGTTILSGI-EATVDPSTQVVYSESPDSGVLADKYDYA 1882
G Q GGWTI+WQG+ T+IL GI +A D V E +G K D A
Sbjct: 461 GKQSGGWTISWQGTGNRNDEFPGATSILGGIRDAVADAGGSV---EFDVAGQYKTKPDVA 517
Query: 1883 IVVVGEPPYAETFGDNLNLTIPAPGPSVIQSVCGAAK-------CVVVLISGRPLVVEPY 2041
IVV GE PYAE GD L P Q + K V V +SGRP+ V P
Sbjct: 518 IVVFGEEPYAEFQGDVETLEYQ---PDQKQDLALLKKLKDQGIPVVAVFLSGRPMWVNPE 574
Query: 2042 LGDMDALVAAWLPGSEGQGVADVLFGD------YGFTGKLPRTWFKSVDQLPMNVGDAHY 2203
L DA VAAWLPG+EG GVADVLF D + F GKL +W ++ Q +N GDA Y
Sbjct: 575 LNASDAFVAAWLPGTEGGGVADVLFTDKAGKVQHDFAGKLSYSWPRTAAQTTVNRGDADY 634
Query: 2204 DPLFPFGFGLTTK 2242
+PLF +G+GLT K
Sbjct: 635 NPLFAYGYGLTYK 647
>sptr|Q8PLK4|Q8PLK4 Glucan 1,4-beta-glucosidase.
Length = 870
Score = 409 bits (1052), Expect = e-113
Identities = 255/622 (40%), Positives = 340/622 (54%), Gaps = 30/622 (4%)
Frame = +2
Query: 461 KYKDPKQPVAV------RIKDLLGRMTLAEKIGQMTQIERENATADALAKYFIGSVLSGG 622
++ PK P A RI D++ +M++ EK+ Q Q + + T D + KY IGSVL+GG
Sbjct: 60 QWPSPKWPFAQDAALEQRITDVMAKMSVEEKVAQTIQGDIASMTPDDVRKYRIGSVLAGG 119
Query: 623 GSVXXXXXXXXXXX--XMVTEMQKGALSTRLG---IPIIYGIDAVHGHNNVYKATIFPHN 787
S + + ++ T G IPII+GIDAVHG +N+ AT+FPHN
Sbjct: 120 NSDPGGKYNASPAEWLKLADAFYEASMDTSKGGNAIPIIFGIDAVHGQSNIVGATLFPHN 179
Query: 788 VGLGATRDPDLVKRIGEATALEVRATGIPYAFAPCIAVCRDPRWGRCYESYSEDPKVVQS 967
+GLGATR+PDL+K+IGE TA E R TG+ + FAP +AV +D RWGR YE YSE P VV S
Sbjct: 180 IGLGATRNPDLIKKIGEVTAAETRVTGMEWTFAPTVAVPQDDRWGRTYEGYSESPDVVAS 239
Query: 968 LT-SLISGLQGDAPADSAGRPYVGGSKKVAACAKHYVGDGGTHNGINENNTIIDTHGLLS 1144
++ G+QG G P V + KH+VGDGGT +G ++ +T + +
Sbjct: 240 FAGKMVEGVQG-----VPGTPQFLDGSHVISSVKHFVGDGGTTDGKDQGDTKVSEATMRD 294
Query: 1145 IHMPPYYNSIIRGVSTVMVSYSSWNGVKMHANHFLVTDFLKNKLKFRGFVISDWEGIDRI 1324
IH Y +I G +VM S++S+NG KMH N ++TD LK ++ F GFV+ DW G ++
Sbjct: 295 IHAAGYPPAIAAGAQSVMASFNSFNGEKMHGNKVMLTDVLKGRMNFGGFVVGDWNGHGQV 354
Query: 1325 TTPPHANYSYSIEAGVGAGIDMIMVPFRYTEFIDDLTTQVQNKVIPMSRIDDAVYRILRV 1504
+ N A AG+DM M + + V++ I R+DDAV RILRV
Sbjct: 355 KGCTNQN----CPASFIAGVDMAMAADSWKGIYETELAAVKSGQISAERLDDAVRRILRV 410
Query: 1505 KFTMGLFENPYPDSSLAGE----LGKQEHRELAREAVRKSLVLLKNGKSSYAPLLPLPKK 1672
K +GLFE P G LG EHR +AR+AVR+SLVLLKN A +LPL
Sbjct: 411 KVRLGLFEAGKPSKRPLGGKYELLGAPEHRAIARQAVRESLVLLKN----QAGILPL-NP 465
Query: 1673 AGKILVAGSHANDLGNQCGGWTITWQGSS---GNTTAGTTILSGIEATVDPSTQVVYSES 1843
++LV G AND+G Q GGWT+ WQG+ + GTTI G++ + + +E
Sbjct: 466 TRRVLVVGDGANDMGKQSGGWTLNWQGTGTKRSDYPNGTTIWEGLDKQIKAAGG--SAEL 523
Query: 1844 PDSGVLADKYDYAIVVVGEPPYAETFGDNLNLTIPAPGPSVIQSVCGAAKC-----VVVL 2008
G K D A+VV GE PYAE GD L + PG ++ K V V
Sbjct: 524 AVDGAYKTKPDVAVVVFGENPYAEFQGDIATL-LYKPGDESELALIKKLKAEGIPVVAVF 582
Query: 2009 ISGRPLVVEPYLGDMDALVAAWLPGSEGQGVADVLFG------DYGFTGKLPRTWFKSVD 2170
+SGRPL + Y+ DA VAAWLPGSEG+G+ADVL F GKL +W K+
Sbjct: 583 LSGRPLWMNQYINASDAFVAAWLPGSEGEGIADVLLRKADGSVQNDFKGKLSFSWPKTAV 642
Query: 2171 QLPMNVGDAHYDPLFPFGFGLT 2236
Q NVG YDP F FGFGLT
Sbjct: 643 QFANNVGQKDYDPQFKFGFGLT 664
>sptr|Q8P9S3|Q8P9S3 Glucan 1,4-beta-glucosidase.
Length = 870
Score = 409 bits (1050), Expect = e-112
Identities = 254/611 (41%), Positives = 337/611 (55%), Gaps = 25/611 (4%)
Frame = +2
Query: 479 QPVAVRIKDLLGRMTLAEKIGQMTQIERENATADALAKYFIGSVLSGGGSVXXXXXXXXX 658
Q + RI D++ +M++ EK+ Q Q + + T D + KY IGSVL+GG S
Sbjct: 72 QALEQRISDVMAKMSVEEKVAQTIQGDIASMTPDDVRKYRIGSVLAGGNSDPGGKYDAKP 131
Query: 659 XX--XMVTEMQKGALSTRLG---IPIIYGIDAVHGHNNVYKATIFPHNVGLGATRDPDLV 823
+ + ++ T G IPII+GIDAVHG +N+ AT+FPHN+GLGATR+PDL+
Sbjct: 132 AEWLKLADAFYEASMDTSKGGNAIPIIFGIDAVHGQSNIVGATLFPHNIGLGATRNPDLI 191
Query: 824 KRIGEATALEVRATGIPYAFAPCIAVCRDPRWGRCYESYSEDPKVVQSLT-SLISGLQGD 1000
K+IGE TA E R TG+ + FAP +AV +D RWGR YE YSE P VV S ++ G+QG
Sbjct: 192 KKIGEVTAAETRVTGMEWTFAPTVAVPQDDRWGRSYEGYSESPDVVASFAGKMVEGVQG- 250
Query: 1001 APADSAGRPYVGGSKKVAACAKHYVGDGGTHNGINENNTIIDTHGLLSIHMPPYYNSIIR 1180
G P V + KH+VGDGGT +G ++ +T + + IH Y +I
Sbjct: 251 ----VPGTPQFLDGSHVISSVKHFVGDGGTTDGKDQGDTKVSEATMRDIHAAGYPPAIAA 306
Query: 1181 GVSTVMVSYSSWNGVKMHANHFLVTDFLKNKLKFRGFVISDWEGIDRITTPPHANYSYSI 1360
G TVM S++S+NG KMH N ++TD LK ++ F GFV+ DW G ++ + N
Sbjct: 307 GAQTVMASFNSFNGEKMHGNKVMLTDVLKGRMNFGGFVVGDWNGHGQVKGCTNEN----C 362
Query: 1361 EAGVGAGIDMIMVPFRYTEFIDDLTTQVQNKVIPMSRIDDAVYRILRVKFTMGLFENPYP 1540
A AG+DM M + + V++ I M R+DDAV RILRVK +GL E P
Sbjct: 363 PASFIAGVDMAMASDSWKGIYETELAAVKSGQISMERLDDAVRRILRVKLRLGLLEAGKP 422
Query: 1541 DSSLAGE----LGKQEHRELAREAVRKSLVLLKNGKSSYAPLLPL-PKKAGKILVAGSHA 1705
G LG EHR +AR+AVR+SLVLLKN + +LPL PKK ++LV G A
Sbjct: 423 SKRPLGGKFELLGAPEHRAIARQAVRESLVLLKN----QSGVLPLDPKK--RVLVVGDGA 476
Query: 1706 NDLGNQCGGWTITWQGSS---GNTTAGTTILSGIEATVDPSTQVVYSESPDSGVLADKYD 1876
ND+G Q GGWT+ WQG+ + G TI G+ + + +E G K D
Sbjct: 477 NDMGKQSGGWTLNWQGTGTKRSDYPNGNTIWEGLNKQITAAGG--SAELAVDGAYKTKPD 534
Query: 1877 YAIVVVGEPPYAETFGDNLNLTIPAPGPSVIQSVCGAAKC-----VVVLISGRPLVVEPY 2041
A+VV GE PYAE GD L + PG ++ K V V +SGRPL + Y
Sbjct: 535 VAVVVFGENPYAEFQGDIATL-LYKPGDDSELALLKKFKAEGIPVVAVFLSGRPLWMNQY 593
Query: 2042 LGDMDALVAAWLPGSEGQGVADVLFG------DYGFTGKLPRTWFKSVDQLPMNVGDAHY 2203
+ DA VAAWLPGSEG+G+ADVL F GKL +W K+ Q NVG Y
Sbjct: 594 INVADAFVAAWLPGSEGEGIADVLLRKADGSVQNDFKGKLSFSWPKTAVQFANNVGQKDY 653
Query: 2204 DPLFPFGFGLT 2236
DP F FGFGLT
Sbjct: 654 DPQFKFGFGLT 664
Database: swall
Posted date: Feb 26, 2004 3:00 PM
Number of letters in database: 439,479,560
Number of sequences in database: 1,381,838
Lambda K H
0.318 0.135 0.401
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 2,060,534,800
Number of Sequences: 1381838
Number of extensions: 48290068
Number of successful extensions: 178001
Number of sequences better than 10.0: 30
Number of HSP's better than 10.0 without gapping: 144888
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 176132
length of database: 439,479,560
effective HSP length: 132
effective length of database: 257,076,944
effective search space used: 189465707728
frameshift window, decay const: 50, 0.1
T: 12
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)

