BLASTX 2.2.6 [Apr-09-2003]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= QBTB.063F13F020916.3.1
(668 letters)
Database: swall
1,381,838 sequences; 439,479,560 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
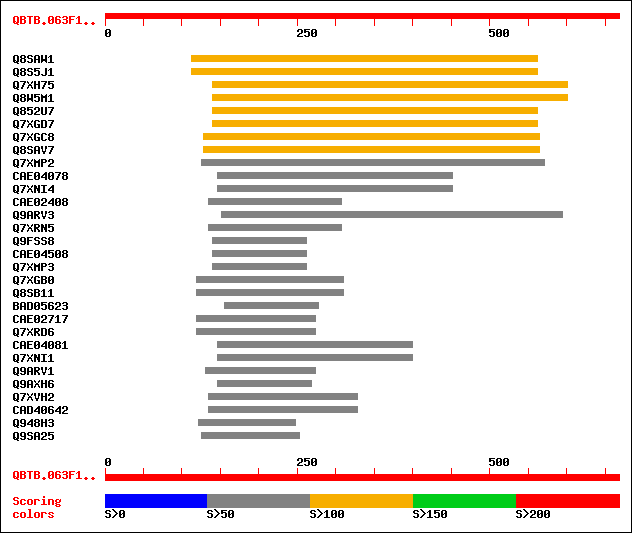
sptr|Q8SAW1|Q8SAW1 Putative wall-associated protein kinase. 123 2e-27
sptr|Q8S5J1|Q8S5J1 Putative wall-associated protein kinase. 123 2e-27
sptr|Q7XH75|Q7XH75 Putative wall-associated protein kinase. 122 6e-27
sptr|Q8W5M1|Q8W5M1 Putative wall-associated protein kinase. 122 6e-27
sptr|Q852U7|Q852U7 Hypothetical protein similar to wall-associat... 120 1e-26
sptr|Q7XGD7|Q7XGD7 Putative wall-associated protein kinase. 120 1e-26
sptr|Q7XGC8|Q7XGC8 Putative wall-associated protein kinase. 111 7e-24
sptr|Q8SAV7|Q8SAV7 Putative wall-associated protein kinase. 111 7e-24
sptr|Q7XMP2|Q7XMP2 OSJNBb0059K02.19 protein. 69 4e-11
sptrnew|CAE04078|CAE04078 OSJNBb0032D24.8 protein. 64 2e-09
sptr|Q7XNI4|Q7XNI4 OSJNBb0032D24.9 protein. 64 2e-09
sptrnew|CAE02408|CAE02408 OSJNBa0024J22.12 protein. 64 2e-09
sptr|Q9ARV3|Q9ARV3 Putative wall-associated kinase 2. 64 2e-09
sptr|Q7XRN5|Q7XRN5 OSJNBa0024J22.13 protein. 64 2e-09
sptr|Q9FSS8|Q9FSS8 Putative wall-associated kinase 2. 63 4e-09
sptrnew|CAE04508|CAE04508 OSJNBb0059K02.18 protein. 63 4e-09
sptr|Q7XMP3|Q7XMP3 OSJNBb0059K02.18 protein. 63 4e-09
sptr|Q7XGB0|Q7XGB0 Putative wall-associated kinase 1. 62 9e-09
sptr|Q8SB11|Q8SB11 Putative wall-associated kinase 1. 62 9e-09
sptrnew|BAD05623|BAD05623 Wall-associated kinase 2-like. 61 1e-08
sptrnew|CAE02717|CAE02717 OSJNBa0055H05.4 protein. 61 1e-08
sptr|Q7XRD6|Q7XRD6 OSJNBa0055H05.6 protein. 61 1e-08
sptrnew|CAE04081|CAE04081 OSJNBb0032D24.11 protein. 61 2e-08
sptr|Q7XNI1|Q7XNI1 OSJNBb0032D24.12 protein. 61 2e-08
sptr|Q9ARV1|Q9ARV1 P0503E05.27 protein (P0697D09.14 protein). 60 3e-08
sptr|Q9AXH6|Q9AXH6 Wall-associated protein kinase. 58 1e-07
sptr|Q7XVH2|Q7XVH2 OSJNBa0016N04.4 protein. 55 1e-06
sptrnew|CAD40642|CAD40642 OSJNBa0016N04.2 protein. 55 1e-06
sptr|Q948H3|Q948H3 Putative wall-associated kinase 2. 52 5e-06
sptr|Q9SA25|Q9SA25 F3O9.6 protein. 52 5e-06
>sptr|Q8SAW1|Q8SAW1 Putative wall-associated protein kinase.
Length = 388
Score = 123 bits (309), Expect = 2e-27
Identities = 69/156 (44%), Positives = 93/156 (59%), Gaps = 6/156 (3%)
Frame = +2
Query: 113 QHSCASPPLPVTLPGCPDKCGNISIPYPFGTKEG-CFKPDFEVLCNDSFHPPRAFIQMAG 289
QH PP +TLPGC DKCGNISIP+PFGTK+ CF P FEV CND+F PPR F+ +
Sbjct: 27 QHVATLPP--ITLPGCIDKCGNISIPFPFGTKQSSCFLPGFEVTCNDTFSPPRLFLGNSN 84
Query: 290 AKIT-----VDDTYYKTNEDGIPDNDIGSSEWPIELVDVSVSEGKARVLGAFSYDCKMND 454
++ YY T E+G+P + + +EL+ ++++EG AR G S DC +ND
Sbjct: 85 PGDRQNYQEFEERYYLTTEEGMPTHLMTDDFLFMELMSINLTEGVARAYGPVSSDCSLND 144
Query: 455 TYHAVRRQEIDLGYWMPFSMSDADDVLMGIGSNVVA 562
TYH V+RQ L PF +S + L +G N+ A
Sbjct: 145 TYHLVKRQMTGLA--GPFLIS-TRNALTAVGWNMEA 177
>sptr|Q8S5J1|Q8S5J1 Putative wall-associated protein kinase.
Length = 706
Score = 123 bits (309), Expect = 2e-27
Identities = 69/156 (44%), Positives = 93/156 (59%), Gaps = 6/156 (3%)
Frame = +2
Query: 113 QHSCASPPLPVTLPGCPDKCGNISIPYPFGTKEG-CFKPDFEVLCNDSFHPPRAFIQMAG 289
QH PP +TLPGC DKCGNISIP+PFGTK+ CF P FEV CND+F PPR F+ +
Sbjct: 27 QHVATLPP--ITLPGCIDKCGNISIPFPFGTKQSSCFLPGFEVTCNDTFSPPRLFLGNSN 84
Query: 290 AKIT-----VDDTYYKTNEDGIPDNDIGSSEWPIELVDVSVSEGKARVLGAFSYDCKMND 454
++ YY T E+G+P + + +EL+ ++++EG AR G S DC +ND
Sbjct: 85 PGDRQNYQEFEERYYLTTEEGMPTHLMTDDFLFMELMSINLTEGVARAYGPVSSDCSLND 144
Query: 455 TYHAVRRQEIDLGYWMPFSMSDADDVLMGIGSNVVA 562
TYH V+RQ L PF +S + L +G N+ A
Sbjct: 145 TYHLVKRQMTGLA--GPFLIS-TRNALTAVGWNMEA 177
>sptr|Q7XH75|Q7XH75 Putative wall-associated protein kinase.
Length = 722
Score = 122 bits (305), Expect = 6e-27
Identities = 69/158 (43%), Positives = 90/158 (56%), Gaps = 4/158 (2%)
Frame = +2
Query: 140 PVTLPGCPDKCGNISIPYPFGTKEGCFKPDFEVLCNDSFHPPRAFIQMAGAKITVD---D 310
P+TL GCPD+CG++SIPYPFG GCF FEV CN +F PPRAF+ D
Sbjct: 29 PITLAGCPDRCGDVSIPYPFGMAPGCFLDGFEVTCNRTFDPPRAFLAWGSPDSPFQGNAD 88
Query: 311 TYYKTNEDGIPDNDIGSSEWPIELVDVSVSEGKARVLGAFSYDC-KMNDTYHAVRRQEID 487
YY ++ D + D S P+ELVDV++S G+AR GA + DC N TYH RRQ
Sbjct: 89 GYYLSDNDSVTLKDYWS--LPVELVDVTLSRGEARAYGAVTTDCAATNGTYHEFRRQLTV 146
Query: 488 LGYWMPFSMSDADDVLMGIGSNVVAKQTSGPQCQSYLL 601
L PF S + +VL G+G ++ A+ T+ Y L
Sbjct: 147 LS-GSPFVFSASRNVLTGVGWDMEAQLTTSLASTGYRL 183
>sptr|Q8W5M1|Q8W5M1 Putative wall-associated protein kinase.
Length = 722
Score = 122 bits (305), Expect = 6e-27
Identities = 69/158 (43%), Positives = 90/158 (56%), Gaps = 4/158 (2%)
Frame = +2
Query: 140 PVTLPGCPDKCGNISIPYPFGTKEGCFKPDFEVLCNDSFHPPRAFIQMAGAKITVD---D 310
P+TL GCPD+CG++SIPYPFG GCF FEV CN +F PPRAF+ D
Sbjct: 29 PITLAGCPDRCGDVSIPYPFGMAPGCFLDGFEVTCNRTFDPPRAFLAWGSPDSPFQGNAD 88
Query: 311 TYYKTNEDGIPDNDIGSSEWPIELVDVSVSEGKARVLGAFSYDC-KMNDTYHAVRRQEID 487
YY ++ D + D S P+ELVDV++S G+AR GA + DC N TYH RRQ
Sbjct: 89 GYYLSDNDSVTLKDYWS--LPVELVDVTLSRGEARAYGAVTTDCAATNGTYHEFRRQLTV 146
Query: 488 LGYWMPFSMSDADDVLMGIGSNVVAKQTSGPQCQSYLL 601
L PF S + +VL G+G ++ A+ T+ Y L
Sbjct: 147 LS-GSPFVFSASRNVLTGVGWDMEAQLTTSLASTGYRL 183
>sptr|Q852U7|Q852U7 Hypothetical protein similar to wall-associated
kinases.
Length = 364
Score = 120 bits (302), Expect = 1e-26
Identities = 62/158 (39%), Positives = 93/158 (58%), Gaps = 17/158 (10%)
Frame = +2
Query: 140 PVTLPGCP--DKCGNISIPYPFGTKEGCFKPDFEVLCNDSFHPPRAFIQ----MAGAKIT 301
P L GCP D CGNI+IP+PFG K GC+ FEV+C+ +F+PPRAF+ + G K+
Sbjct: 25 PTELQGCPAPDMCGNITIPHPFGIKPGCYLAGFEVICDRTFNPPRAFLAGDPPLFGDKLP 84
Query: 302 VD-----------DTYYKTNEDGIPDNDIGSSEWPIELVDVSVSEGKARVLGAFSYDCKM 448
+ + YY + G+P + + P+EL+D+SV++ K RV A + DC
Sbjct: 85 PNVNSSKPFTVTANFYYSGTDAGMPSEMVNYTRGPLELLDISVNQSKLRVYAAINSDCST 144
Query: 449 NDTYHAVRRQEIDLGYWMPFSMSDADDVLMGIGSNVVA 562
N+T+H + Q I L PF++S D+ L+G+G NV+A
Sbjct: 145 NETHHILFEQSIKLQPSGPFTLSANDNSLVGVGQNVIA 182
>sptr|Q7XGD7|Q7XGD7 Putative wall-associated protein kinase.
Length = 360
Score = 120 bits (302), Expect = 1e-26
Identities = 62/158 (39%), Positives = 93/158 (58%), Gaps = 17/158 (10%)
Frame = +2
Query: 140 PVTLPGCP--DKCGNISIPYPFGTKEGCFKPDFEVLCNDSFHPPRAFIQ----MAGAKIT 301
P L GCP D CGNI+IP+PFG K GC+ FEV+C+ +F+PPRAF+ + G K+
Sbjct: 25 PTELQGCPAPDMCGNITIPHPFGIKPGCYLAGFEVICDRTFNPPRAFLAGDPPLFGDKLP 84
Query: 302 VD-----------DTYYKTNEDGIPDNDIGSSEWPIELVDVSVSEGKARVLGAFSYDCKM 448
+ + YY + G+P + + P+EL+D+SV++ K RV A + DC
Sbjct: 85 PNVNSSKPFTVTANFYYSGTDAGMPSEMVNYTRGPLELLDISVNQSKLRVYAAINSDCST 144
Query: 449 NDTYHAVRRQEIDLGYWMPFSMSDADDVLMGIGSNVVA 562
N+T+H + Q I L PF++S D+ L+G+G NV+A
Sbjct: 145 NETHHILFEQSIKLQPSGPFTLSANDNSLVGVGQNVIA 182
>sptr|Q7XGC8|Q7XGC8 Putative wall-associated protein kinase.
Length = 338
Score = 111 bits (278), Expect = 7e-24
Identities = 60/147 (40%), Positives = 83/147 (56%), Gaps = 1/147 (0%)
Frame = +2
Query: 128 SPPLPVTLPGCPDKCGNISIPYPFGTKEGCFKP-DFEVLCNDSFHPPRAFIQMAGAKITV 304
SP + LPGCPDKCG+ISIPYPFGTKEGC+ +F VLCN S PP AG I
Sbjct: 34 SPRRRIALPGCPDKCGDISIPYPFGTKEGCYLDINFIVLCNLSTTPPAT---AAGTIILK 90
Query: 305 DDTYYKTNEDGIPDNDIGSSEWPIELVDVSVSEGKARVLGAFSYDCKMNDTYHAVRRQEI 484
+ YY +++ S W ++L+D+ V+ G+ RV S DC N++YH +
Sbjct: 91 GNGYYFGDQENPVGVQTNKSWWTVDLIDIDVTRGEVRVAVPVSSDCSTNESYHELSIFTQ 150
Query: 485 DLGYWMPFSMSDADDVLMGIGSNVVAK 565
L + F S +VL+G+G +V A+
Sbjct: 151 SLNFSTTFLFSATRNVLLGVGQSVRAR 177
>sptr|Q8SAV7|Q8SAV7 Putative wall-associated protein kinase.
Length = 338
Score = 111 bits (278), Expect = 7e-24
Identities = 60/147 (40%), Positives = 83/147 (56%), Gaps = 1/147 (0%)
Frame = +2
Query: 128 SPPLPVTLPGCPDKCGNISIPYPFGTKEGCFKP-DFEVLCNDSFHPPRAFIQMAGAKITV 304
SP + LPGCPDKCG+ISIPYPFGTKEGC+ +F VLCN S PP AG I
Sbjct: 34 SPRRRIALPGCPDKCGDISIPYPFGTKEGCYLDINFIVLCNLSTTPPAT---AAGTIILK 90
Query: 305 DDTYYKTNEDGIPDNDIGSSEWPIELVDVSVSEGKARVLGAFSYDCKMNDTYHAVRRQEI 484
+ YY +++ S W ++L+D+ V+ G+ RV S DC N++YH +
Sbjct: 91 GNGYYFGDQENPVGVQTNKSWWTVDLIDIDVTRGEVRVAVPVSSDCSTNESYHELSIFTQ 150
Query: 485 DLGYWMPFSMSDADDVLMGIGSNVVAK 565
L + F S +VL+G+G +V A+
Sbjct: 151 SLNFSTTFLFSATRNVLLGVGQSVRAR 177
>sptr|Q7XMP2|Q7XMP2 OSJNBb0059K02.19 protein.
Length = 783
Score = 69.3 bits (168), Expect = 4e-11
Identities = 42/155 (27%), Positives = 67/155 (43%), Gaps = 6/155 (3%)
Frame = +2
Query: 125 ASPPLPVTLPGCPDKCGNISIPYPFGTKEGCFKPDFEVLCNDSFH----PPRAFIQMAGA 292
A+ P P+ LPGCP+ CG I +PYPFG +GC F + C+D H PP+ F+
Sbjct: 53 AAAPTPIALPGCPESCGGIQVPYPFGIGDGCSYHGFNLTCDDEAHHHQTPPKLFMATDNG 112
Query: 293 KITVDDTYYKTNEDGIPDNDIGSSEWPIELVDVSVSEGKARVLGAFSYD--CKMNDTYHA 466
+ ++++++S+ +G RV S + +
Sbjct: 113 TV-------------------------VQVLNISLPDGTVRVRSKLSQSSIAGSSSSSSN 147
Query: 467 VRRQEIDLGYWMPFSMSDADDVLMGIGSNVVAKQT 571
DL PF++S A + L+ G N+VA T
Sbjct: 148 ASSSRSDLPADGPFTVSSAYNWLVAFGCNIVADLT 182
>sptrnew|CAE04078|CAE04078 OSJNBb0032D24.8 protein.
Length = 849
Score = 63.9 bits (154), Expect = 2e-09
Identities = 37/114 (32%), Positives = 55/114 (48%), Gaps = 12/114 (10%)
Frame = +2
Query: 146 TLPGCPDKCGNISIPYPFGTKEGCFK-PDFEVLCNDSFHPPRAFIQMAGAKITVD-DTYY 319
TL CP CG +SI YPFG GCF+ PDF ++C++S PP+ + ++ D D+
Sbjct: 3 TLASCPKSCGQMSIHYPFGIGAGCFRQPDFNLICDNSTQPPKLLLHDGTTEVVGDEDSLR 62
Query: 320 KT----------NEDGIPDNDIGSSEWPIELVDVSVSEGKARVLGAFSYDCKMN 451
+T D D+G SEW +D+++S V G Y+ N
Sbjct: 63 RTRYPVRGVRIWRSDSSIGMDVGLSEW----IDINISATIPMVPGVAEYNYSWN 112
Score = 37.7 bits (86), Expect = 0.14
Identities = 16/32 (50%), Positives = 22/32 (68%), Gaps = 1/32 (3%)
Frame = +2
Query: 158 CPDKCGNISIPYPFGTKEGC-FKPDFEVLCND 250
C CGNIS+P+PFG +EGC + FE+ C +
Sbjct: 310 CSRWCGNISVPFPFGLEEGCTARKLFELNCTN 341
>sptr|Q7XNI4|Q7XNI4 OSJNBb0032D24.9 protein.
Length = 849
Score = 63.9 bits (154), Expect = 2e-09
Identities = 37/114 (32%), Positives = 55/114 (48%), Gaps = 12/114 (10%)
Frame = +2
Query: 146 TLPGCPDKCGNISIPYPFGTKEGCFK-PDFEVLCNDSFHPPRAFIQMAGAKITVD-DTYY 319
TL CP CG +SI YPFG GCF+ PDF ++C++S PP+ + ++ D D+
Sbjct: 3 TLASCPKSCGQMSIHYPFGIGAGCFRQPDFNLICDNSTQPPKLLLHDGTTEVVGDEDSLR 62
Query: 320 KT----------NEDGIPDNDIGSSEWPIELVDVSVSEGKARVLGAFSYDCKMN 451
+T D D+G SEW +D+++S V G Y+ N
Sbjct: 63 RTRYPVRGVRIWRSDSSIGMDVGLSEW----IDINISATIPMVPGVAEYNYSWN 112
Score = 37.7 bits (86), Expect = 0.14
Identities = 16/32 (50%), Positives = 22/32 (68%), Gaps = 1/32 (3%)
Frame = +2
Query: 158 CPDKCGNISIPYPFGTKEGC-FKPDFEVLCND 250
C CGNIS+P+PFG +EGC + FE+ C +
Sbjct: 310 CSRWCGNISVPFPFGLEEGCTARKLFELNCTN 341
>sptrnew|CAE02408|CAE02408 OSJNBa0024J22.12 protein.
Length = 937
Score = 63.5 bits (153), Expect = 2e-09
Identities = 28/59 (47%), Positives = 37/59 (62%), Gaps = 1/59 (1%)
Frame = +2
Query: 134 PLPVTLPGCPDKCGNISIPYPFGTKEGCFK-PDFEVLCNDSFHPPRAFIQMAGAKITVD 307
P TL GCP CGN+S YPFG GCF+ PDF + C+++ PPR F+Q G ++ D
Sbjct: 49 PSAATLEGCPRSCGNLSFDYPFGIGSGCFRNPDFNLTCDNTAQPPRLFLQ-GGTEVIED 106
Score = 40.8 bits (94), Expect = 0.016
Identities = 16/33 (48%), Positives = 23/33 (69%), Gaps = 1/33 (3%)
Frame = +2
Query: 158 CPDKCGNISIPYPFGTKEGCF-KPDFEVLCNDS 253
C CGNIS+P+PFG +EGCF + F++ C +
Sbjct: 324 CSRSCGNISVPFPFGLEEGCFARKLFQLNCTSA 356
>sptr|Q9ARV3|Q9ARV3 Putative wall-associated kinase 2.
Length = 659
Score = 63.5 bits (153), Expect = 2e-09
Identities = 48/150 (32%), Positives = 69/150 (46%), Gaps = 2/150 (1%)
Frame = +2
Query: 152 PGCPDKCGNISIPYPFGTKEGC-FKPDFEVLCNDSFHPPRAFIQMAGAKITVDDTYYKTN 328
P C +CG++ IPYPFG GC FE++CN N
Sbjct: 13 PWCKKQCGDVKIPYPFGIGTGCAIGEGFEIICN-------------------------RN 47
Query: 329 EDGIPDNDIGSSEWPIELVDVSVSEGKARVLGAFSYDCKMNDTYHA-VRRQEIDLGYWMP 505
DGI G+ IE++D+SV G++RVLG+ + +C + T A V +DL P
Sbjct: 48 ADGIDQPFTGN----IEVLDISVVYGRSRVLGSITTNCYNSSTGSANVNSWWMDLS-TSP 102
Query: 506 FSMSDADDVLMGIGSNVVAKQTSGPQCQSY 595
+ SDA + + IG N +A +G SY
Sbjct: 103 YRFSDAYNTFVVIGCNTLAYIYNGLNRTSY 132
>sptr|Q7XRN5|Q7XRN5 OSJNBa0024J22.13 protein.
Length = 831
Score = 63.5 bits (153), Expect = 2e-09
Identities = 28/59 (47%), Positives = 37/59 (62%), Gaps = 1/59 (1%)
Frame = +2
Query: 134 PLPVTLPGCPDKCGNISIPYPFGTKEGCFK-PDFEVLCNDSFHPPRAFIQMAGAKITVD 307
P TL GCP CGN+S YPFG GCF+ PDF + C+++ PPR F+Q G ++ D
Sbjct: 41 PSAATLEGCPRSCGNLSFDYPFGIGSGCFRNPDFNLTCDNTAQPPRLFLQ-GGTEVIED 98
Score = 40.8 bits (94), Expect = 0.016
Identities = 16/33 (48%), Positives = 23/33 (69%), Gaps = 1/33 (3%)
Frame = +2
Query: 158 CPDKCGNISIPYPFGTKEGCF-KPDFEVLCNDS 253
C CGNIS+P+PFG +EGCF + F++ C +
Sbjct: 316 CSRSCGNISVPFPFGLEEGCFARKLFQLNCTSA 348
>sptr|Q9FSS8|Q9FSS8 Putative wall-associated kinase 2.
Length = 490
Score = 62.8 bits (151), Expect = 4e-09
Identities = 24/41 (58%), Positives = 30/41 (73%)
Frame = +2
Query: 140 PVTLPGCPDKCGNISIPYPFGTKEGCFKPDFEVLCNDSFHP 262
PV LPGCP+ CGN+++PYPFG GCF+ FE+ C D HP
Sbjct: 21 PVALPGCPETCGNVTVPYPFGIGHGCFRDGFELAC-DETHP 60
>sptrnew|CAE04508|CAE04508 OSJNBb0059K02.18 protein.
Length = 315
Score = 62.8 bits (151), Expect = 4e-09
Identities = 24/41 (58%), Positives = 30/41 (73%)
Frame = +2
Query: 140 PVTLPGCPDKCGNISIPYPFGTKEGCFKPDFEVLCNDSFHP 262
PV LPGCP+ CGN+++PYPFG GCF+ FE+ C D HP
Sbjct: 21 PVALPGCPETCGNVTVPYPFGIGHGCFRDGFELAC-DETHP 60
>sptr|Q7XMP3|Q7XMP3 OSJNBb0059K02.18 protein.
Length = 318
Score = 62.8 bits (151), Expect = 4e-09
Identities = 24/41 (58%), Positives = 30/41 (73%)
Frame = +2
Query: 140 PVTLPGCPDKCGNISIPYPFGTKEGCFKPDFEVLCNDSFHP 262
PV LPGCP+ CGN+++PYPFG GCF+ FE+ C D HP
Sbjct: 21 PVALPGCPETCGNVTVPYPFGIGHGCFRDGFELAC-DETHP 60
>sptr|Q7XGB0|Q7XGB0 Putative wall-associated kinase 1.
Length = 1023
Score = 61.6 bits (148), Expect = 9e-09
Identities = 27/65 (41%), Positives = 39/65 (60%), Gaps = 1/65 (1%)
Frame = +2
Query: 119 SCASPPLPVTLPGCPDKCGNISIPYPFGTKEGCFK-PDFEVLCNDSFHPPRAFIQMAGAK 295
S AS P TL CP +CGN+S YPFG +GCF+ PDF + CN + PP+ + + +
Sbjct: 65 SNASLPSAATLANCPKRCGNLSFDYPFGIGDGCFRHPDFSLTCNATTQPPKLLLHINESV 124
Query: 296 ITVDD 310
+D+
Sbjct: 125 EVIDN 129
Score = 37.7 bits (86), Expect = 0.14
Identities = 14/32 (43%), Positives = 23/32 (71%), Gaps = 1/32 (3%)
Frame = +2
Query: 158 CPDKCGNISIPYPFGTKEGC-FKPDFEVLCND 250
C +CG I++P+PFG +EGC + F++ C+D
Sbjct: 400 CARQCGTITVPFPFGLEEGCSARKRFQLNCSD 431
>sptr|Q8SB11|Q8SB11 Putative wall-associated kinase 1.
Length = 1023
Score = 61.6 bits (148), Expect = 9e-09
Identities = 27/65 (41%), Positives = 39/65 (60%), Gaps = 1/65 (1%)
Frame = +2
Query: 119 SCASPPLPVTLPGCPDKCGNISIPYPFGTKEGCFK-PDFEVLCNDSFHPPRAFIQMAGAK 295
S AS P TL CP +CGN+S YPFG +GCF+ PDF + CN + PP+ + + +
Sbjct: 65 SNASLPSAATLANCPKRCGNLSFDYPFGIGDGCFRHPDFSLTCNATTQPPKLLLHINESV 124
Query: 296 ITVDD 310
+D+
Sbjct: 125 EVIDN 129
Score = 37.7 bits (86), Expect = 0.14
Identities = 14/32 (43%), Positives = 23/32 (71%), Gaps = 1/32 (3%)
Frame = +2
Query: 158 CPDKCGNISIPYPFGTKEGC-FKPDFEVLCND 250
C +CG I++P+PFG +EGC + F++ C+D
Sbjct: 400 CARQCGTITVPFPFGLEEGCSARKRFQLNCSD 431
>sptrnew|BAD05623|BAD05623 Wall-associated kinase 2-like.
Length = 370
Score = 61.2 bits (147), Expect = 1e-08
Identities = 22/41 (53%), Positives = 28/41 (68%)
Frame = +2
Query: 155 GCPDKCGNISIPYPFGTKEGCFKPDFEVLCNDSFHPPRAFI 277
GC CGNIS+PYPFG + C+ P F + CN S+HPP F+
Sbjct: 32 GCTRSCGNISVPYPFGVEADCYYPGFNLTCNHSYHPPTLFL 72
>sptrnew|CAE02717|CAE02717 OSJNBa0055H05.4 protein.
Length = 351
Score = 61.2 bits (147), Expect = 1e-08
Identities = 29/67 (43%), Positives = 34/67 (50%), Gaps = 15/67 (22%)
Frame = +2
Query: 119 SCASPP----------LPVTLPGCPDKCGNISIPYPFGTKEGCF-----KPDFEVLCNDS 253
SCASPP PV CPDKCGN+SIPYPFG GC+ F + CN
Sbjct: 37 SCASPPSPSPSPPPLIFPVPASNCPDKCGNVSIPYPFGIGRGCYLDLPGSGSFSITCNHK 96
Query: 254 FHPPRAF 274
PP+ +
Sbjct: 97 TDPPQPY 103
>sptr|Q7XRD6|Q7XRD6 OSJNBa0055H05.6 protein.
Length = 465
Score = 61.2 bits (147), Expect = 1e-08
Identities = 29/67 (43%), Positives = 34/67 (50%), Gaps = 15/67 (22%)
Frame = +2
Query: 119 SCASPP----------LPVTLPGCPDKCGNISIPYPFGTKEGCF-----KPDFEVLCNDS 253
SCASPP PV CPDKCGN+SIPYPFG GC+ F + CN
Sbjct: 37 SCASPPSPSPSPPPLIFPVPASNCPDKCGNVSIPYPFGIGRGCYLDLPGSGSFSITCNHK 96
Query: 254 FHPPRAF 274
PP+ +
Sbjct: 97 TDPPQPY 103
>sptrnew|CAE04081|CAE04081 OSJNBb0032D24.11 protein.
Length = 802
Score = 60.8 bits (146), Expect = 2e-08
Identities = 34/86 (39%), Positives = 48/86 (55%), Gaps = 1/86 (1%)
Frame = +2
Query: 146 TLPGCPDKCGNISIPYPFGTKEGCFK-PDFEVLCNDSFHPPRAFIQMAGAKITVDDTYYK 322
TL CP CG +SI YPFG GCF+ PDF ++C++S PP+ + G + DT
Sbjct: 3 TLASCPKSCGQMSIHYPFGIGAGCFRQPDFNLICDNSTQPPKLLLH-DGVTEIIGDT--- 58
Query: 323 TNEDGIPDNDIGSSEWPIELVDVSVS 400
D D D+GS+ E +DV++S
Sbjct: 59 ---DSSFDMDVGST----ESIDVNIS 77
Score = 38.9 bits (89), Expect = 0.062
Identities = 13/20 (65%), Positives = 17/20 (85%)
Frame = +2
Query: 158 CPDKCGNISIPYPFGTKEGC 217
C +CGNIS+P+PFG +EGC
Sbjct: 270 CSRRCGNISVPFPFGLEEGC 289
>sptr|Q7XNI1|Q7XNI1 OSJNBb0032D24.12 protein.
Length = 802
Score = 60.8 bits (146), Expect = 2e-08
Identities = 34/86 (39%), Positives = 48/86 (55%), Gaps = 1/86 (1%)
Frame = +2
Query: 146 TLPGCPDKCGNISIPYPFGTKEGCFK-PDFEVLCNDSFHPPRAFIQMAGAKITVDDTYYK 322
TL CP CG +SI YPFG GCF+ PDF ++C++S PP+ + G + DT
Sbjct: 3 TLASCPKSCGQMSIHYPFGIGAGCFRQPDFNLICDNSTQPPKLLLH-DGVTEIIGDT--- 58
Query: 323 TNEDGIPDNDIGSSEWPIELVDVSVS 400
D D D+GS+ E +DV++S
Sbjct: 59 ---DSSFDMDVGST----ESIDVNIS 77
Score = 38.9 bits (89), Expect = 0.062
Identities = 13/20 (65%), Positives = 17/20 (85%)
Frame = +2
Query: 158 CPDKCGNISIPYPFGTKEGC 217
C +CGNIS+P+PFG +EGC
Sbjct: 270 CSRRCGNISVPFPFGLEEGC 289
>sptr|Q9ARV1|Q9ARV1 P0503E05.27 protein (P0697D09.14 protein).
Length = 189
Score = 59.7 bits (143), Expect = 3e-08
Identities = 23/48 (47%), Positives = 31/48 (64%)
Frame = +2
Query: 131 PPLPVTLPGCPDKCGNISIPYPFGTKEGCFKPDFEVLCNDSFHPPRAF 274
PP L CP CG+++I YPFG GCF+P FE++CN + PP+ F
Sbjct: 24 PPSARDLRHCPTSCGDVNITYPFGIGTGCFRPGFELICNTTTKPPKLF 71
>sptr|Q9AXH6|Q9AXH6 Wall-associated protein kinase.
Length = 722
Score = 57.8 bits (138), Expect = 1e-07
Identities = 22/41 (53%), Positives = 29/41 (70%)
Frame = +2
Query: 146 TLPGCPDKCGNISIPYPFGTKEGCFKPDFEVLCNDSFHPPR 268
T+ GCP KCG++ IP PFG + C F+V+CN+SF PPR
Sbjct: 35 TVAGCPSKCGDVDIPLPFGIGDHCAWESFDVVCNESFSPPR 75
>sptr|Q7XVH2|Q7XVH2 OSJNBa0016N04.4 protein.
Length = 381
Score = 54.7 bits (130), Expect = 1e-06
Identities = 27/71 (38%), Positives = 38/71 (53%), Gaps = 6/71 (8%)
Frame = +2
Query: 134 PLPVTLPGCPDKCGNISIPYPFGTKEGCFK-PDFEVLCNDSFHPPR-----AFIQMAGAK 295
P TL GC CG+++ YPFG GCF+ PDFE+ C+ + PPR Q+AG+
Sbjct: 45 PSTATLAGCQSSCGDLTFVYPFGIGSGCFRSPDFELTCDSTTSPPRLLFHDGITQIAGSI 104
Query: 296 ITVDDTYYKTN 328
V + T+
Sbjct: 105 NIVSTEFMDTD 115
>sptrnew|CAD40642|CAD40642 OSJNBa0016N04.2 protein.
Length = 455
Score = 54.7 bits (130), Expect = 1e-06
Identities = 27/71 (38%), Positives = 38/71 (53%), Gaps = 6/71 (8%)
Frame = +2
Query: 134 PLPVTLPGCPDKCGNISIPYPFGTKEGCFK-PDFEVLCNDSFHPPR-----AFIQMAGAK 295
P TL GC CG+++ YPFG GCF+ PDFE+ C+ + PPR Q+AG+
Sbjct: 45 PSTATLAGCQSSCGDLTFVYPFGIGSGCFRSPDFELTCDSTTSPPRLLFHDGITQIAGSI 104
Query: 296 ITVDDTYYKTN 328
V + T+
Sbjct: 105 NIVSTEFMDTD 115
Score = 40.4 bits (93), Expect = 0.021
Identities = 17/33 (51%), Positives = 23/33 (69%), Gaps = 1/33 (3%)
Frame = +2
Query: 158 CPDKCGNISIPYPFGTKEGCF-KPDFEVLCNDS 253
C +CGNIS+P+PFG +EGCF F + C +S
Sbjct: 328 CIRQCGNISVPFPFGLEEGCFATKGFYLNCTNS 360
>sptr|Q948H3|Q948H3 Putative wall-associated kinase 2.
Length = 769
Score = 52.4 bits (124), Expect = 5e-06
Identities = 24/46 (52%), Positives = 30/46 (65%), Gaps = 4/46 (8%)
Frame = +2
Query: 122 CASPPLPVTL---PGCPDKCGNISIPYPFGTKEGCFK-PDFEVLCN 247
C +P P + PGCPDKCGNISI YPFG GC + DF++ C+
Sbjct: 21 CLAPVTPASAQPWPGCPDKCGNISISYPFGIGAGCARDKDFQLECD 66
>sptr|Q9SA25|Q9SA25 F3O9.6 protein.
Length = 720
Score = 52.4 bits (124), Expect = 5e-06
Identities = 21/44 (47%), Positives = 29/44 (65%), Gaps = 1/44 (2%)
Frame = +2
Query: 125 ASPPLPVTLPGCPDKCGNISIPYPFGTKEGCFKPD-FEVLCNDS 253
A+ P+ L C D CGN+S+PYPFG +GC+K FE++C S
Sbjct: 22 AASTFPLALRNCSDHCGNVSVPYPFGIGKGCYKNKWFEIVCKSS 65
Database: swall
Posted date: Feb 26, 2004 3:00 PM
Number of letters in database: 439,479,560
Number of sequences in database: 1,381,838
Lambda K H
0.318 0.135 0.401
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 476,017,511
Number of Sequences: 1381838
Number of extensions: 9793552
Number of successful extensions: 34153
Number of sequences better than 10.0: 30
Number of HSP's better than 10.0 without gapping: 32856
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 34093
length of database: 439,479,560
effective HSP length: 119
effective length of database: 275,040,838
effective search space used: 28329206314
frameshift window, decay const: 50, 0.1
T: 12
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)

