BLASTX 2.2.6 [Apr-09-2003]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
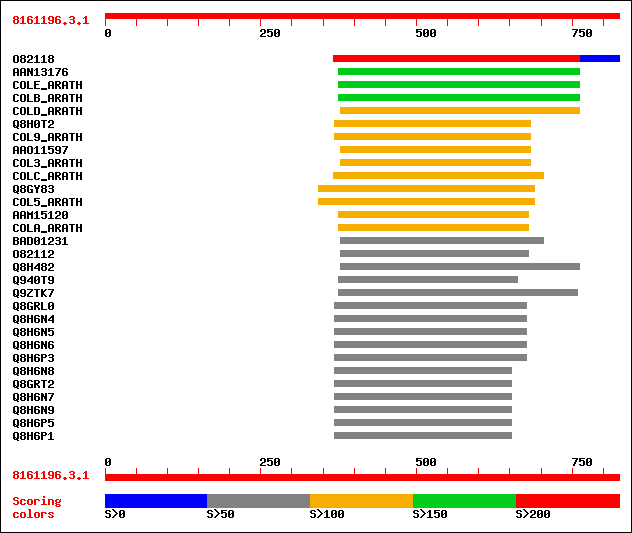
Query= 8161196.3.1
(825 letters)
Database: swall
1,381,838 sequences; 439,479,560 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
sptr|O82118|O82118 Zinc finger protein. 234 4e-63
sptrnew|AAN13176|AAN13176 Hypothetical protein. 192 6e-48
sw|Q9LUA9|COLE_ARATH Zinc finger protein constans-like 14. 192 6e-48
sw|Q9SSE5|COLB_ARATH Zinc finger protein constans-like 11. 181 1e-44
sw|O23379|COLD_ARATH Hypothetical zinc finger protein constans-l... 132 5e-30
sptr|Q8H0T2|Q8H0T2 Putative CONSTANS-like B-box zinc finger prot... 131 1e-29
sw|O22800|COL9_ARATH Zinc finger protein constans-like 9. 131 1e-29
sptrnew|AAO11597|AAO11597 At1g28050/F13K9_15. 131 1e-29
sw|Q9C7E8|COL3_ARATH Zinc finger protein constans-like 3. 131 1e-29
sw|Q9LJ44|COLC_ARATH Zinc finger protein constans-like 12. 127 1e-28
sptr|Q8GY83|Q8GY83 Putative zinc finger protein. 114 1e-24
sw|Q9C9F4|COL5_ARATH Zinc finger protein constans-like 5. 114 1e-24
sptrnew|AAM15120|AAM15120 Putative zinc-finger protein (B-box zi... 107 3e-22
sw|O82256|COLA_ARATH Zinc finger protein constans-like 10. 107 3e-22
sptrnew|BAD01231|BAD01231 Zinc-finger protein C60910-like. 92 7e-18
sptr|O82112|O82112 Zinc finger protein. 86 6e-16
sptr|Q8H482|Q8H482 Putative zinc-finger protein. 86 8e-16
sptr|Q940T9|Q940T9 AT5g24930/F6A4_140. 84 3e-15
sptr|Q9ZTK7|Q9ZTK7 CONSTANS-like protein 2. 84 3e-15
sptr|Q8GRL0|Q8GRL0 COL1 protein. 83 4e-15
sptr|Q8H6N4|Q8H6N4 COL1 protein. 83 4e-15
sptr|Q8H6N5|Q8H6N5 COL1 protein. 83 4e-15
sptr|Q8H6N6|Q8H6N6 COL1 protein. 83 4e-15
sptr|Q8H6P3|Q8H6P3 COL1 protein. 83 4e-15
sptr|Q8H6N8|Q8H6N8 COL1 protein. 82 7e-15
sptr|Q8GRT2|Q8GRT2 COL1 protein. 82 7e-15
sptr|Q8H6N7|Q8H6N7 COL1 protein. 82 7e-15
sptr|Q8H6N9|Q8H6N9 COL1 protein. 82 7e-15
sptr|Q8H6P5|Q8H6P5 COL1 protein. 82 7e-15
sptr|Q8H6P1|Q8H6P1 COL1 protein. 82 7e-15
>sptr|O82118|O82118 Zinc finger protein.
Length = 407
Score = 234 bits (597), Expect(2) = 4e-63
Identities = 108/132 (81%), Positives = 119/132 (90%)
Frame = +3
Query: 366 MVPLCGFCGKQRSMIYCRSDAASLCLSCDRSVHSANALSRRHRRTLLCDRCGLQPASVRC 545
M LC FC +QRSM+YCRSDAASLCLSCDR+VHSANALSRRH RTLLCDRC QPA+VRC
Sbjct: 1 MDALCDFCREQRSMVYCRSDAASLCLSCDRNVHSANALSRRHTRTLLCDRCVGQPAAVRC 60
Query: 546 LEDNTSLCQNCDWNGHDAASGASGHKRQAINCYSGCPSSAELSRIWSFITDIPTVAAEPN 725
LE+NTSLCQNCDWNGH AAS A+GHKRQ INCYSGCPSSAELSRIWSF DIPTVAAEPN
Sbjct: 61 LEENTSLCQNCDWNGHGAASSAAGHKRQTINCYSGCPSSAELSRIWSFSMDIPTVAAEPN 120
Query: 726 YEDGLSMMTIDD 761
E+G++MM+I+D
Sbjct: 121 CEEGINMMSIND 132
Score = 30.0 bits (66), Expect(2) = 4e-63
Identities = 13/21 (61%), Positives = 17/21 (80%)
Frame = +1
Query: 763 SDVTNHHGASDDKRLLEIANT 825
+DV NH GA +D RLL+IA+T
Sbjct: 133 NDVNNHCGAPEDGRLLDIAST 153
>sptrnew|AAN13176|AAN13176 Hypothetical protein.
Length = 373
Score = 192 bits (487), Expect = 6e-48
Identities = 88/132 (66%), Positives = 106/132 (80%), Gaps = 3/132 (2%)
Frame = +3
Query: 375 LCGFCGKQRSMIYCRSDAASLCLSCDRSVHSANALSRRHRRTLLCDRCGLQPASVRCLED 554
+C FCG+QRSM+YCRSDAA LCLSCDR+VHSANALS+RH RTL+C+RC QPASVRC ++
Sbjct: 4 MCDFCGEQRSMVYCRSDAACLCLSCDRNVHSANALSKRHSRTLVCERCNAQPASVRCSDE 63
Query: 555 NTSLCQNCDWNGHDA--ASGASGHKRQAINCYSGCPSSAELSRIWSFITDIPTVAAEPN- 725
SLCQNCDW+GHD ++ S HKRQ INCYSGCPSSAELS IWSF D+ +AE +
Sbjct: 64 RVSLCQNCDWSGHDGKNSTTTSHHKRQTINCYSGCPSSAELSSIWSFCMDLNISSAEESA 123
Query: 726 YEDGLSMMTIDD 761
E G+ +MTID+
Sbjct: 124 CEQGMGLMTIDE 135
>sw|Q9LUA9|COLE_ARATH Zinc finger protein constans-like 14.
Length = 373
Score = 192 bits (487), Expect = 6e-48
Identities = 88/132 (66%), Positives = 106/132 (80%), Gaps = 3/132 (2%)
Frame = +3
Query: 375 LCGFCGKQRSMIYCRSDAASLCLSCDRSVHSANALSRRHRRTLLCDRCGLQPASVRCLED 554
+C FCG+QRSM+YCRSDAA LCLSCDR+VHSANALS+RH RTL+C+RC QPASVRC ++
Sbjct: 4 MCDFCGEQRSMVYCRSDAACLCLSCDRNVHSANALSKRHSRTLVCERCNAQPASVRCSDE 63
Query: 555 NTSLCQNCDWNGHDA--ASGASGHKRQAINCYSGCPSSAELSRIWSFITDIPTVAAEPN- 725
SLCQNCDW+GHD ++ S HKRQ INCYSGCPSSAELS IWSF D+ +AE +
Sbjct: 64 RVSLCQNCDWSGHDGKNSTTTSHHKRQTINCYSGCPSSAELSSIWSFCMDLNISSAEESA 123
Query: 726 YEDGLSMMTIDD 761
E G+ +MTID+
Sbjct: 124 CEQGMGLMTIDE 135
>sw|Q9SSE5|COLB_ARATH Zinc finger protein constans-like 11.
Length = 372
Score = 181 bits (459), Expect = 1e-44
Identities = 84/138 (60%), Positives = 103/138 (74%), Gaps = 9/138 (6%)
Frame = +3
Query: 375 LCGFCGKQRSMIYCRSDAASLCLSCDRSVHSANALSRRHRRTLLCDRCGLQPASVRCLED 554
+C FCG+QRSM+YCRSDAA LCLSCDRSVHSANALS+RH RTL+C+RC QPA+VRC+E+
Sbjct: 4 MCDFCGEQRSMVYCRSDAACLCLSCDRSVHSANALSKRHSRTLVCERCNAQPATVRCVEE 63
Query: 555 NTSLCQNCDWNGHD---------AASGASGHKRQAINCYSGCPSSAELSRIWSFITDIPT 707
SLCQNCDW+GH+ +++ HKRQ I+CYSGCPSS+EL+ IWSF D+
Sbjct: 64 RVSLCQNCDWSGHNNSNNNNSSSSSTSPQQHKRQTISCYSGCPSSSELASIWSFCLDL-- 121
Query: 708 VAAEPNYEDGLSMMTIDD 761
A + E L MM IDD
Sbjct: 122 -AGQSICEQELGMMNIDD 138
>sw|O23379|COLD_ARATH Hypothetical zinc finger protein constans-like
13.
Length = 330
Score = 132 bits (333), Expect = 5e-30
Identities = 57/128 (44%), Positives = 84/128 (65%)
Frame = +3
Query: 378 CGFCGKQRSMIYCRSDAASLCLSCDRSVHSANALSRRHRRTLLCDRCGLQPASVRCLEDN 557
C FCG ++++IYC+SD+A LCL+CD +VHSAN LS+RH R+LLC++C LQP +V C+ +N
Sbjct: 5 CDFCGTEKALIYCKSDSAKLCLNCDVNVHSANPLSQRHTRSLLCEKCSLQPTAVHCMNEN 64
Query: 558 TSLCQNCDWNGHDAASGASGHKRQAINCYSGCPSSAELSRIWSFITDIPTVAAEPNYEDG 737
SLCQ C W + GH+ Q++N YS CPS ++ +IWS + + + D
Sbjct: 65 VSLCQGCQWTASNCT--GLGHRLQSLNPYSDCPSPSDFGKIWSSTLEPSVTSLVSPFSDT 122
Query: 738 LSMMTIDD 761
L + +DD
Sbjct: 123 L-LQELDD 129
>sptr|Q8H0T2|Q8H0T2 Putative CONSTANS-like B-box zinc finger
protein.
Length = 402
Score = 131 bits (330), Expect = 1e-29
Identities = 53/105 (50%), Positives = 74/105 (70%)
Frame = +3
Query: 369 VPLCGFCGKQRSMIYCRSDAASLCLSCDRSVHSANALSRRHRRTLLCDRCGLQPASVRCL 548
V C FCG++ ++++CR+D A LCL CD+ VHSAN LSR+H R+ +CD C +P SVRC
Sbjct: 9 VVACEFCGERTAVLFCRADTAKLCLPCDQHVHSANLLSRKHVRSQICDNCSKEPVSVRCF 68
Query: 549 EDNTSLCQNCDWNGHDAASGASGHKRQAINCYSGCPSSAELSRIW 683
DN LCQ CDW+ H + S ++ H+R A+ +SGCPS EL+ +W
Sbjct: 69 TDNLVLCQECDWDVHGSCSSSATHERSAVEGFSGCPSVLELAAVW 113
>sw|O22800|COL9_ARATH Zinc finger protein constans-like 9.
Length = 361
Score = 131 bits (330), Expect = 1e-29
Identities = 53/105 (50%), Positives = 74/105 (70%)
Frame = +3
Query: 369 VPLCGFCGKQRSMIYCRSDAASLCLSCDRSVHSANALSRRHRRTLLCDRCGLQPASVRCL 548
V C FCG++ ++++CR+D A LCL CD+ VHSAN LSR+H R+ +CD C +P SVRC
Sbjct: 9 VVACEFCGERTAVLFCRADTAKLCLPCDQHVHSANLLSRKHVRSQICDNCSKEPVSVRCF 68
Query: 549 EDNTSLCQNCDWNGHDAASGASGHKRQAINCYSGCPSSAELSRIW 683
DN LCQ CDW+ H + S ++ H+R A+ +SGCPS EL+ +W
Sbjct: 69 TDNLVLCQECDWDVHGSCSSSATHERSAVEGFSGCPSVLELAAVW 113
>sptrnew|AAO11597|AAO11597 At1g28050/F13K9_15.
Length = 433
Score = 131 bits (330), Expect = 1e-29
Identities = 52/102 (50%), Positives = 73/102 (71%)
Frame = +3
Query: 378 CGFCGKQRSMIYCRSDAASLCLSCDRSVHSANALSRRHRRTLLCDRCGLQPASVRCLEDN 557
C FCG++ ++++CR+D A LCL CD+ VH+AN LSR+H R+ +CD CG +P SVRC DN
Sbjct: 9 CDFCGERTAVLFCRADTAKLCLPCDQQVHTANLLSRKHVRSQICDNCGNEPVSVRCFTDN 68
Query: 558 TSLCQNCDWNGHDAASGASGHKRQAINCYSGCPSSAELSRIW 683
LCQ CDW+ H + S + H R A+ +SGCPS+ EL+ +W
Sbjct: 69 LILCQECDWDVHGSCSVSDAHVRSAVEGFSGCPSALELAALW 110
>sw|Q9C7E8|COL3_ARATH Zinc finger protein constans-like 3.
Length = 433
Score = 131 bits (330), Expect = 1e-29
Identities = 52/102 (50%), Positives = 73/102 (71%)
Frame = +3
Query: 378 CGFCGKQRSMIYCRSDAASLCLSCDRSVHSANALSRRHRRTLLCDRCGLQPASVRCLEDN 557
C FCG++ ++++CR+D A LCL CD+ VH+AN LSR+H R+ +CD CG +P SVRC DN
Sbjct: 9 CDFCGERTAVLFCRADTAKLCLPCDQQVHTANLLSRKHVRSQICDNCGNEPVSVRCFTDN 68
Query: 558 TSLCQNCDWNGHDAASGASGHKRQAINCYSGCPSSAELSRIW 683
LCQ CDW+ H + S + H R A+ +SGCPS+ EL+ +W
Sbjct: 69 LILCQECDWDVHGSCSVSDAHVRSAVEGFSGCPSALELAALW 110
>sw|Q9LJ44|COLC_ARATH Zinc finger protein constans-like 12.
Length = 337
Score = 127 bits (320), Expect = 1e-28
Identities = 54/113 (47%), Positives = 77/113 (68%)
Frame = +3
Query: 366 MVPLCGFCGKQRSMIYCRSDAASLCLSCDRSVHSANALSRRHRRTLLCDRCGLQPASVRC 545
M P C C +++IYC+SD A LCL+CD VHSAN LS RH R+L+C++C QPA++RC
Sbjct: 1 MEPKCDHCATSQALIYCKSDLAKLCLNCDVHVHSANPLSHRHIRSLICEKCFSQPAAIRC 60
Query: 546 LEDNTSLCQNCDWNGHDAASGASGHKRQAINCYSGCPSSAELSRIWSFITDIP 704
L++ S CQ C W H++ GH+ Q++N +SGCPS + +R+WS I + P
Sbjct: 61 LDEKVSYCQGCHW--HESNCSELGHRVQSLNPFSGCPSPTDFNRMWSSILEPP 111
>sptr|Q8GY83|Q8GY83 Putative zinc finger protein.
Length = 356
Score = 114 bits (286), Expect = 1e-24
Identities = 50/116 (43%), Positives = 73/116 (62%)
Frame = +3
Query: 342 ISLILSRIMVPLCGFCGKQRSMIYCRSDAASLCLSCDRSVHSANALSRRHRRTLLCDRCG 521
+ +I+ M +C FC R+++YC +D A+LCL+CD VHSAN+LS RH RT+LCD C
Sbjct: 2 LCIIIIENMERVCEFCKAYRAVVYCIADTANLCLTCDAKVHSANSLSGRHLRTVLCDSCK 61
Query: 522 LQPASVRCLEDNTSLCQNCDWNGHDAASGASGHKRQAINCYSGCPSSAELSRIWSF 689
QP VRC + LC C+ H G+S H+R+ + CY+GCP + + + +W F
Sbjct: 62 NQPCVVRCFDHKMFLCHGCNDKFH--GGGSSEHRRRDLRCYTGCPPAKDFAVMWGF 115
>sw|Q9C9F4|COL5_ARATH Zinc finger protein constans-like 5.
Length = 356
Score = 114 bits (286), Expect = 1e-24
Identities = 50/116 (43%), Positives = 73/116 (62%)
Frame = +3
Query: 342 ISLILSRIMVPLCGFCGKQRSMIYCRSDAASLCLSCDRSVHSANALSRRHRRTLLCDRCG 521
+ +I+ M +C FC R+++YC +D A+LCL+CD VHSAN+LS RH RT+LCD C
Sbjct: 2 LCIIIIENMERVCEFCKAYRAVVYCIADTANLCLTCDAKVHSANSLSGRHLRTVLCDSCK 61
Query: 522 LQPASVRCLEDNTSLCQNCDWNGHDAASGASGHKRQAINCYSGCPSSAELSRIWSF 689
QP VRC + LC C+ H G+S H+R+ + CY+GCP + + + +W F
Sbjct: 62 NQPCVVRCFDHKMFLCHGCNDKFH--GGGSSEHRRRDLRCYTGCPPAKDFAVMWGF 115
>sptrnew|AAM15120|AAM15120 Putative zinc-finger protein (B-box zinc
finger domain).
Length = 332
Score = 107 bits (266), Expect = 3e-22
Identities = 47/102 (46%), Positives = 66/102 (64%)
Frame = +3
Query: 375 LCGFCGKQRSMIYCRSDAASLCLSCDRSVHSANALSRRHRRTLLCDRCGLQPASVRCLED 554
LC +C +++YC++D+A LCL+CD+ VH AN L +H R+LLCD C P+S+ C +
Sbjct: 12 LCDYCDSSVALVYCKADSAKLCLACDKQVHVANQLFAKHFRSLLCDSCNESPSSLFCETE 71
Query: 555 NTSLCQNCDWNGHDAASGASGHKRQAINCYSGCPSSAELSRI 680
+ LCQNCDW H A+S S H R+ ++GCPS EL I
Sbjct: 72 RSVLCQNCDWQHHTASS--SLHSRRPFEGFTGCPSVPELLAI 111
>sw|O82256|COLA_ARATH Zinc finger protein constans-like 10.
Length = 332
Score = 107 bits (266), Expect = 3e-22
Identities = 47/102 (46%), Positives = 66/102 (64%)
Frame = +3
Query: 375 LCGFCGKQRSMIYCRSDAASLCLSCDRSVHSANALSRRHRRTLLCDRCGLQPASVRCLED 554
LC +C +++YC++D+A LCL+CD+ VH AN L +H R+LLCD C P+S+ C +
Sbjct: 12 LCDYCDSSVALVYCKADSAKLCLACDKQVHVANQLFAKHFRSLLCDSCNESPSSLFCETE 71
Query: 555 NTSLCQNCDWNGHDAASGASGHKRQAINCYSGCPSSAELSRI 680
+ LCQNCDW H A+S S H R+ ++GCPS EL I
Sbjct: 72 RSVLCQNCDWQHHTASS--SLHSRRPFEGFTGCPSVPELLAI 111
>sptrnew|BAD01231|BAD01231 Zinc-finger protein C60910-like.
Length = 488
Score = 92.4 bits (228), Expect = 7e-18
Identities = 47/111 (42%), Positives = 68/111 (61%), Gaps = 2/111 (1%)
Frame = +3
Query: 378 CGFCGKQRSMIYCRSDAASLCLSCDRSVHSANALSRRHRRTLLCDRCGLQPASVRCLEDN 557
C +CG+ + ++CR+DAA LC++CDR VH+ANALSR+H R LC C +PA+ R +
Sbjct: 18 CDYCGEAAAALHCRADAARLCVACDRHVHAANALSRKHVRAPLCAACAARPAAARVASAS 77
Query: 558 TS--LCQNCDWNGHDAASGASGHKRQAINCYSGCPSSAELSRIWSFITDIP 704
LC +CD G GA+ R + +SGCP++AEL+ W D+P
Sbjct: 78 APAFLCADCD-TGCGGDDGAA--LRVPVEGFSGCPAAAELAASWGL--DLP 123
>sptr|O82112|O82112 Zinc finger protein.
Length = 403
Score = 85.9 bits (211), Expect = 6e-16
Identities = 42/105 (40%), Positives = 58/105 (55%), Gaps = 4/105 (3%)
Frame = +3
Query: 378 CGFCGKQRSMIYCRSDAASLCLSCDRSVHSANALSRRHRRTLLCDRCGLQPASVRCLEDN 557
C +CG +++YCR+DAA LCL CDR VH AN + RH R LC C A R
Sbjct: 38 CDYCGDAAAVVYCRADAARLCLPCDRHVHGANGVCSRHARAPLCAACAAAGAVFR-RGAG 96
Query: 558 TSLCQNCDWNGH----DAASGASGHKRQAINCYSGCPSSAELSRI 680
LC NCD++ H + A H R ++ Y+GCPS+ +L+ +
Sbjct: 97 GFLCSNCDFSRHRHGGERDPAAPLHDRSTVHPYTGCPSALDLAAL 141
>sptr|Q8H482|Q8H482 Putative zinc-finger protein.
Length = 379
Score = 85.5 bits (210), Expect = 8e-16
Identities = 49/141 (34%), Positives = 73/141 (51%), Gaps = 13/141 (9%)
Frame = +3
Query: 378 CGFCGKQRSMIYCRSDAASLCLSCDRSVHSANALSRRHRRTLLCDRCGLQPASVRCLEDN 557
C FCG +++YCR+D+A LCL CDR VH+AN +S RH R LC C PA+ D
Sbjct: 24 CDFCGGLPAVVYCRADSARLCLPCDRHVHAANTVSTRHARAPLCSACRAAPAAAFHRGDG 83
Query: 558 TSLCQNCDWNG--HDAASGASGHK-----RQAINCYSGCPSSAELSRIWSFITDIPTVAA 716
LC +CD++ + G G + R A+ Y+GCPS EL+ I + A
Sbjct: 84 -FLCSSCDFDERLRRGSIGGGGDELPLDDRAAVEGYTGCPSIGELAAILGVVGGDSDKPA 142
Query: 717 EPNY------EDGLSMMTIDD 761
+ + E+ ++++DD
Sbjct: 143 DDGWWSASWEEEAPQVLSLDD 163
>sptr|Q940T9|Q940T9 AT5g24930/F6A4_140.
Length = 232
Score = 83.6 bits (205), Expect = 3e-15
Identities = 41/96 (42%), Positives = 52/96 (54%)
Frame = +3
Query: 375 LCGFCGKQRSMIYCRSDAASLCLSCDRSVHSANALSRRHRRTLLCDRCGLQPASVRCLED 554
LC C + +YCR DAA LCLSCD VH+AN L+ RH R +C+ C PA V C D
Sbjct: 5 LCDSCKSATAALYCRPDAAFLCLSCDSKVHAANKLASRHARVWMCEVCEQAPAHVTCKAD 64
Query: 555 NTSLCQNCDWNGHDAASGASGHKRQAINCYSGCPSS 662
+LC CD + H A A H+R + + SS
Sbjct: 65 AAALCVTCDRDIHSANPLARRHERVPVTPFYDSVSS 100
>sptr|Q9ZTK7|Q9ZTK7 CONSTANS-like protein 2.
Length = 329
Score = 83.6 bits (205), Expect = 3e-15
Identities = 43/128 (33%), Positives = 66/128 (51%)
Frame = +3
Query: 375 LCGFCGKQRSMIYCRSDAASLCLSCDRSVHSANALSRRHRRTLLCDRCGLQPASVRCLED 554
LC C + ++CR+D+A LC++CD +H+AN L+ RH R LC+ C PA V C D
Sbjct: 5 LCDSCQSATATLFCRADSAFLCVNCDSKIHAANKLASRHPRVWLCEVCEQAPAHVTCKAD 64
Query: 555 NTSLCQNCDWNGHDAASGASGHKRQAINCYSGCPSSAELSRIWSFITDIPTVAAEPNYED 734
+ +LC CD + H A +S H R + + +SA S +P V + N+ D
Sbjct: 65 DAALCVTCDRDIHSANPLSSRHDRVPVTPFYDSVNSAANS--------VPVVKSVVNFLD 116
Query: 735 GLSMMTID 758
+ +D
Sbjct: 117 DRYLSDVD 124
>sptr|Q8GRL0|Q8GRL0 COL1 protein.
Length = 345
Score = 83.2 bits (204), Expect = 4e-15
Identities = 40/100 (40%), Positives = 53/100 (53%)
Frame = +3
Query: 378 CGFCGKQRSMIYCRSDAASLCLSCDRSVHSANALSRRHRRTLLCDRCGLQPASVRCLEDN 557
C C +YCR+D+A LC SCD +H+AN L+ RH R +C+ C PA+ C D
Sbjct: 12 CDTCRSAACTVYCRADSAYLCTSCDAQIHAANRLASRHERVRVCESCERAPAAFFCKADA 71
Query: 558 TSLCQNCDWNGHDAASGASGHKRQAINCYSGCPSSAELSR 677
SLC CD H A A H+R I SGC ++ S+
Sbjct: 72 ASLCTACDSQIHSANPLARRHQRVPILPISGCVATNHSSK 111
Score = 53.5 bits (127), Expect = 4e-06
Identities = 21/43 (48%), Positives = 31/43 (72%)
Frame = +3
Query: 369 VPLCGFCGKQRSMIYCRSDAASLCLSCDRSVHSANALSRRHRR 497
V +C C + + +C++DAASLC +CD +HSAN L+RRH+R
Sbjct: 52 VRVCESCERAPAAFFCKADAASLCTACDSQIHSANPLARRHQR 94
>sptr|Q8H6N4|Q8H6N4 COL1 protein.
Length = 345
Score = 83.2 bits (204), Expect = 4e-15
Identities = 40/100 (40%), Positives = 53/100 (53%)
Frame = +3
Query: 378 CGFCGKQRSMIYCRSDAASLCLSCDRSVHSANALSRRHRRTLLCDRCGLQPASVRCLEDN 557
C C +YCR+D+A LC SCD +H+AN L+ RH R +C+ C PA+ C D
Sbjct: 12 CDTCRSAACTVYCRADSAYLCTSCDAQIHAANRLASRHERVRVCESCERAPAAFFCKADA 71
Query: 558 TSLCQNCDWNGHDAASGASGHKRQAINCYSGCPSSAELSR 677
SLC CD H A A H+R I SGC ++ S+
Sbjct: 72 ASLCTACDSQIHSANPLARRHQRVPILPISGCVATNHSSK 111
Score = 53.5 bits (127), Expect = 4e-06
Identities = 21/43 (48%), Positives = 31/43 (72%)
Frame = +3
Query: 369 VPLCGFCGKQRSMIYCRSDAASLCLSCDRSVHSANALSRRHRR 497
V +C C + + +C++DAASLC +CD +HSAN L+RRH+R
Sbjct: 52 VRVCESCERAPAAFFCKADAASLCTACDSQIHSANPLARRHQR 94
>sptr|Q8H6N5|Q8H6N5 COL1 protein.
Length = 345
Score = 83.2 bits (204), Expect = 4e-15
Identities = 40/100 (40%), Positives = 53/100 (53%)
Frame = +3
Query: 378 CGFCGKQRSMIYCRSDAASLCLSCDRSVHSANALSRRHRRTLLCDRCGLQPASVRCLEDN 557
C C +YCR+D+A LC SCD +H+AN L+ RH R +C+ C PA+ C D
Sbjct: 12 CDTCRSAACTVYCRADSAYLCTSCDAQIHAANRLASRHERVRVCESCERAPAAFFCKADA 71
Query: 558 TSLCQNCDWNGHDAASGASGHKRQAINCYSGCPSSAELSR 677
SLC CD H A A H+R I SGC ++ S+
Sbjct: 72 ASLCTACDSQIHSANPLARRHQRVPILPISGCVATNHSSK 111
Score = 53.5 bits (127), Expect = 4e-06
Identities = 21/43 (48%), Positives = 31/43 (72%)
Frame = +3
Query: 369 VPLCGFCGKQRSMIYCRSDAASLCLSCDRSVHSANALSRRHRR 497
V +C C + + +C++DAASLC +CD +HSAN L+RRH+R
Sbjct: 52 VRVCESCERAPAAFFCKADAASLCTACDSQIHSANPLARRHQR 94
>sptr|Q8H6N6|Q8H6N6 COL1 protein.
Length = 345
Score = 83.2 bits (204), Expect = 4e-15
Identities = 40/100 (40%), Positives = 53/100 (53%)
Frame = +3
Query: 378 CGFCGKQRSMIYCRSDAASLCLSCDRSVHSANALSRRHRRTLLCDRCGLQPASVRCLEDN 557
C C +YCR+D+A LC SCD +H+AN L+ RH R +C+ C PA+ C D
Sbjct: 12 CDTCRSAACTVYCRADSAYLCTSCDAQIHAANRLASRHERVRVCESCERAPAAFFCKADA 71
Query: 558 TSLCQNCDWNGHDAASGASGHKRQAINCYSGCPSSAELSR 677
SLC CD H A A H+R I SGC ++ S+
Sbjct: 72 ASLCTACDSQIHSANPLARRHQRVPILPISGCVATNHSSK 111
Score = 53.5 bits (127), Expect = 4e-06
Identities = 21/43 (48%), Positives = 31/43 (72%)
Frame = +3
Query: 369 VPLCGFCGKQRSMIYCRSDAASLCLSCDRSVHSANALSRRHRR 497
V +C C + + +C++DAASLC +CD +HSAN L+RRH+R
Sbjct: 52 VRVCESCERAPAAFFCKADAASLCTACDSQIHSANPLARRHQR 94
>sptr|Q8H6P3|Q8H6P3 COL1 protein.
Length = 345
Score = 83.2 bits (204), Expect = 4e-15
Identities = 40/100 (40%), Positives = 53/100 (53%)
Frame = +3
Query: 378 CGFCGKQRSMIYCRSDAASLCLSCDRSVHSANALSRRHRRTLLCDRCGLQPASVRCLEDN 557
C C +YCR+D+A LC SCD +H+AN L+ RH R +C+ C PA+ C D
Sbjct: 12 CDTCRSAACTVYCRADSAYLCTSCDAQIHAANRLASRHERVRVCESCERAPAAFFCKADA 71
Query: 558 TSLCQNCDWNGHDAASGASGHKRQAINCYSGCPSSAELSR 677
SLC CD H A A H+R I SGC ++ S+
Sbjct: 72 ASLCTACDSQIHSANPLARRHQRVPILPISGCVATNHSSK 111
Score = 53.5 bits (127), Expect = 4e-06
Identities = 21/43 (48%), Positives = 31/43 (72%)
Frame = +3
Query: 369 VPLCGFCGKQRSMIYCRSDAASLCLSCDRSVHSANALSRRHRR 497
V +C C + + +C++DAASLC +CD +HSAN L+RRH+R
Sbjct: 52 VRVCESCERAPAAFFCKADAASLCTACDSQIHSANPLARRHQR 94
>sptr|Q8H6N8|Q8H6N8 COL1 protein.
Length = 344
Score = 82.4 bits (202), Expect = 7e-15
Identities = 39/92 (42%), Positives = 49/92 (53%)
Frame = +3
Query: 378 CGFCGKQRSMIYCRSDAASLCLSCDRSVHSANALSRRHRRTLLCDRCGLQPASVRCLEDN 557
C C +YCR+D+A LC SCD +H+AN L+ RH R +C+ C PA+ C D
Sbjct: 12 CDTCRSAACTVYCRADSAYLCTSCDAQIHAANRLASRHERVRVCESCERAPAAFFCKADA 71
Query: 558 TSLCQNCDWNGHDAASGASGHKRQAINCYSGC 653
SLC CD H A A H+R I SGC
Sbjct: 72 ASLCTACDSQIHSANPLARRHQRVPILPISGC 103
Score = 53.5 bits (127), Expect = 4e-06
Identities = 21/43 (48%), Positives = 31/43 (72%)
Frame = +3
Query: 369 VPLCGFCGKQRSMIYCRSDAASLCLSCDRSVHSANALSRRHRR 497
V +C C + + +C++DAASLC +CD +HSAN L+RRH+R
Sbjct: 52 VRVCESCERAPAAFFCKADAASLCTACDSQIHSANPLARRHQR 94
>sptr|Q8GRT2|Q8GRT2 COL1 protein.
Length = 342
Score = 82.4 bits (202), Expect = 7e-15
Identities = 39/92 (42%), Positives = 49/92 (53%)
Frame = +3
Query: 378 CGFCGKQRSMIYCRSDAASLCLSCDRSVHSANALSRRHRRTLLCDRCGLQPASVRCLEDN 557
C C +YCR+D+A LC SCD +H+AN L+ RH R +C+ C PA+ C D
Sbjct: 12 CDTCRSAACTVYCRADSAYLCTSCDAQIHAANRLASRHERVRVCESCERAPAAFFCKADA 71
Query: 558 TSLCQNCDWNGHDAASGASGHKRQAINCYSGC 653
SLC CD H A A H+R I SGC
Sbjct: 72 ASLCTACDSQIHSANPLARRHQRVPILPISGC 103
Score = 53.5 bits (127), Expect = 4e-06
Identities = 21/43 (48%), Positives = 31/43 (72%)
Frame = +3
Query: 369 VPLCGFCGKQRSMIYCRSDAASLCLSCDRSVHSANALSRRHRR 497
V +C C + + +C++DAASLC +CD +HSAN L+RRH+R
Sbjct: 52 VRVCESCERAPAAFFCKADAASLCTACDSQIHSANPLARRHQR 94
>sptr|Q8H6N7|Q8H6N7 COL1 protein.
Length = 342
Score = 82.4 bits (202), Expect = 7e-15
Identities = 39/92 (42%), Positives = 49/92 (53%)
Frame = +3
Query: 378 CGFCGKQRSMIYCRSDAASLCLSCDRSVHSANALSRRHRRTLLCDRCGLQPASVRCLEDN 557
C C +YCR+D+A LC SCD +H+AN L+ RH R +C+ C PA+ C D
Sbjct: 12 CDTCRSAACTVYCRADSAYLCTSCDAQIHAANRLASRHERVRVCESCERAPAAFFCKADA 71
Query: 558 TSLCQNCDWNGHDAASGASGHKRQAINCYSGC 653
SLC CD H A A H+R I SGC
Sbjct: 72 ASLCTACDSQIHSANPLARRHQRVPILPISGC 103
Score = 53.5 bits (127), Expect = 4e-06
Identities = 21/43 (48%), Positives = 31/43 (72%)
Frame = +3
Query: 369 VPLCGFCGKQRSMIYCRSDAASLCLSCDRSVHSANALSRRHRR 497
V +C C + + +C++DAASLC +CD +HSAN L+RRH+R
Sbjct: 52 VRVCESCERAPAAFFCKADAASLCTACDSQIHSANPLARRHQR 94
>sptr|Q8H6N9|Q8H6N9 COL1 protein.
Length = 342
Score = 82.4 bits (202), Expect = 7e-15
Identities = 39/92 (42%), Positives = 49/92 (53%)
Frame = +3
Query: 378 CGFCGKQRSMIYCRSDAASLCLSCDRSVHSANALSRRHRRTLLCDRCGLQPASVRCLEDN 557
C C +YCR+D+A LC SCD +H+AN L+ RH R +C+ C PA+ C D
Sbjct: 12 CDTCRSAACTVYCRADSAYLCTSCDAQIHAANRLASRHERVRVCESCERAPAAFFCKADA 71
Query: 558 TSLCQNCDWNGHDAASGASGHKRQAINCYSGC 653
SLC CD H A A H+R I SGC
Sbjct: 72 ASLCTACDSQIHSANPLARRHQRVPILPISGC 103
Score = 53.5 bits (127), Expect = 4e-06
Identities = 21/43 (48%), Positives = 31/43 (72%)
Frame = +3
Query: 369 VPLCGFCGKQRSMIYCRSDAASLCLSCDRSVHSANALSRRHRR 497
V +C C + + +C++DAASLC +CD +HSAN L+RRH+R
Sbjct: 52 VRVCESCERAPAAFFCKADAASLCTACDSQIHSANPLARRHQR 94
>sptr|Q8H6P5|Q8H6P5 COL1 protein.
Length = 342
Score = 82.4 bits (202), Expect = 7e-15
Identities = 39/92 (42%), Positives = 49/92 (53%)
Frame = +3
Query: 378 CGFCGKQRSMIYCRSDAASLCLSCDRSVHSANALSRRHRRTLLCDRCGLQPASVRCLEDN 557
C C +YCR+D+A LC SCD +H+AN L+ RH R +C+ C PA+ C D
Sbjct: 12 CDTCRSAACTVYCRADSAYLCTSCDAQIHAANRLASRHERVRVCESCERAPAAFFCKADA 71
Query: 558 TSLCQNCDWNGHDAASGASGHKRQAINCYSGC 653
SLC CD H A A H+R I SGC
Sbjct: 72 ASLCTACDSQIHSANPLARRHQRVPILPISGC 103
Score = 53.5 bits (127), Expect = 4e-06
Identities = 21/43 (48%), Positives = 31/43 (72%)
Frame = +3
Query: 369 VPLCGFCGKQRSMIYCRSDAASLCLSCDRSVHSANALSRRHRR 497
V +C C + + +C++DAASLC +CD +HSAN L+RRH+R
Sbjct: 52 VRVCESCERAPAAFFCKADAASLCTACDSQIHSANPLARRHQR 94
>sptr|Q8H6P1|Q8H6P1 COL1 protein.
Length = 345
Score = 82.4 bits (202), Expect = 7e-15
Identities = 39/92 (42%), Positives = 49/92 (53%)
Frame = +3
Query: 378 CGFCGKQRSMIYCRSDAASLCLSCDRSVHSANALSRRHRRTLLCDRCGLQPASVRCLEDN 557
C C +YCR+D+A LC SCD +H+AN L+ RH R +C+ C PA+ C D
Sbjct: 12 CDTCRSAACTVYCRADSAYLCTSCDAQIHAANRLASRHERVRVCESCERAPAAFFCKADA 71
Query: 558 TSLCQNCDWNGHDAASGASGHKRQAINCYSGC 653
SLC CD H A A H+R I SGC
Sbjct: 72 ASLCTACDSQIHSANPLARRHQRVPILPISGC 103
Score = 53.5 bits (127), Expect = 4e-06
Identities = 21/43 (48%), Positives = 31/43 (72%)
Frame = +3
Query: 369 VPLCGFCGKQRSMIYCRSDAASLCLSCDRSVHSANALSRRHRR 497
V +C C + + +C++DAASLC +CD +HSAN L+RRH+R
Sbjct: 52 VRVCESCERAPAAFFCKADAASLCTACDSQIHSANPLARRHQR 94
Database: swall
Posted date: Feb 26, 2004 3:00 PM
Number of letters in database: 439,479,560
Number of sequences in database: 1,381,838
Lambda K H
0.318 0.135 0.401
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 690,936,016
Number of Sequences: 1381838
Number of extensions: 15132330
Number of successful extensions: 53541
Number of sequences better than 10.0: 30
Number of HSP's better than 10.0 without gapping: 48679
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 53237
length of database: 439,479,560
effective HSP length: 121
effective length of database: 272,277,162
effective search space used: 41658405786
frameshift window, decay const: 50, 0.1
T: 12
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)

