BLASTX 2.2.6 [Apr-09-2003]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
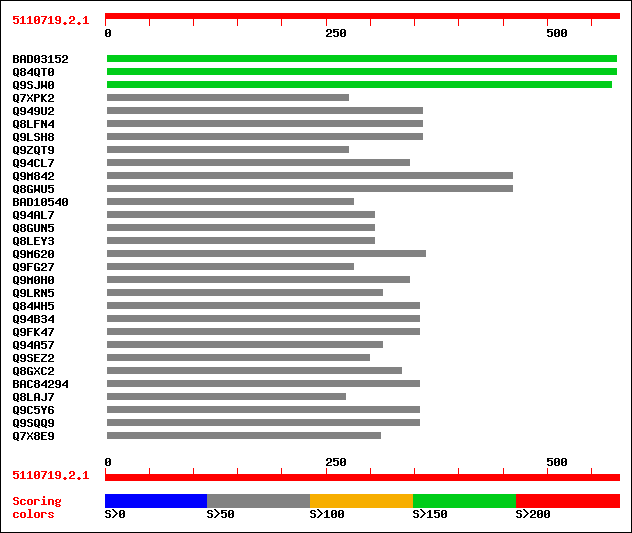
Query= 5110719.2.1
(581 letters)
Database: /db/uniprot/tmp/swall
1,395,590 sequences; 442,889,342 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
sptrnew|BAD03152|BAD03152 Putative transfactor. 199 2e-50
sptr|Q84QT0|Q84QT0 Putative transfactor. 199 2e-50
sptr|Q9SJW0|Q9SJW0 Transfactor-like protein. 154 1e-36
sptr|Q7XPK2|Q7XPK2 OSJNBa0087O24.8 protein. 79 5e-14
sptr|Q949U2|Q949U2 Hypothetical protein. 77 2e-13
sptr|Q8LFN4|Q8LFN4 Transfactor-like protein. 77 2e-13
sptr|Q9LSH8|Q9LSH8 Transfactor-like protein. 77 2e-13
sptr|Q9ZQT9|Q9ZQT9 WERBP-1 protein. 76 3e-13
sptr|Q94CL7|Q94CL7 Phosphate starvation response regulator 1 (Hy... 76 3e-13
sptr|Q9M842|Q9M842 Transfactor, putative. 75 8e-13
sptr|Q8GWU5|Q8GWU5 Putative transfactor (At3g04445). 74 1e-12
sptrnew|BAD10540|BAD10540 Putative transfactor. 74 2e-12
sptr|Q94AL7|Q94AL7 Hypothetical protein (Fragment). 72 5e-12
sptr|Q8GUN5|Q8GUN5 Hypothetical protein. 72 5e-12
sptr|Q8LEY3|Q8LEY3 Transfactor, putative. 72 5e-12
sptr|Q9M620|Q9M620 CDPK substrate protein 1. 70 2e-11
sptr|Q9FG27|Q9FG27 Similarity to transfactor. 69 4e-11
sptr|Q9M0H0|Q9M0H0 Hypothetical protein. 69 4e-11
sptr|Q9LRN5|Q9LRN5 Similarity to transfactor. 67 2e-10
sptr|Q84WH5|Q84WH5 Transfactor-like protein. 65 5e-10
sptr|Q94B34|Q94B34 Transfactor-like protein. 65 5e-10
sptr|Q9FK47|Q9FK47 Transfactor-like protein. 65 5e-10
sptr|Q94A57|Q94A57 AT3g24120/MUJ8_3 (Transfactor, putative). 65 6e-10
sptr|Q9SEZ2|Q9SEZ2 Transfactor, putative, 28697-27224. 64 1e-09
sptr|Q8GXC2|Q8GXC2 Hypothetical protein. 63 2e-09
sptrnew|BAC84294|BAC84294 Putative CDPK substrate protein 1. 63 3e-09
sptr|Q8LAJ7|Q8LAJ7 Transfactor, putative. 62 7e-09
sptr|Q9C5Y6|Q9C5Y6 MYR1. 61 9e-09
sptr|Q9SQQ9|Q9SQQ9 Transfactor-like. 59 3e-08
sptr|Q7X8E9|Q7X8E9 Putative phosphate starvation response regula... 57 1e-07
>sptrnew|BAD03152|BAD03152 Putative transfactor.
Length = 307
Score = 199 bits (506), Expect = 2e-50
Identities = 109/192 (56%), Positives = 125/192 (65%)
Frame = +3
Query: 3 LTIYHVKSHLQKYRLAKYIPDASTDGNKTDNKDPGDLLAGLEGSSGLQISEALKLQMEVQ 182
LTIYHVKSHLQKYRLAKYIPD S D NK ++KDPG+LL+ LEGSSG+QISEALKLQMEVQ
Sbjct: 85 LTIYHVKSHLQKYRLAKYIPDPSADDNKDEDKDPGNLLSALEGSSGMQISEALKLQMEVQ 144
Query: 183 KXXXXXXXXXXXXXXXXXAQGKYLQKIIEEQQRLTGVKSETPAGGASVTVSSDQFPDSER 362
K AQGKYLQKIIEEQQR+ G GAS SS+Q PDSE+
Sbjct: 145 KRLHEQLEVQRQLQLRIEAQGKYLQKIIEEQQRVIG-------AGASRATSSEQLPDSEK 197
Query: 363 XXXXXXXXXXXXXXQVGASNRDTGDRTEATKSTCHGDSLSRNEPLTPDSNCQNGSPVASP 542
Q ++++ + E TKS H D+L EPLTPDS+C+ GSP SP
Sbjct: 198 TNPPTPVPISESPVQGAPHSKNSQSQVEPTKSPSHDDALPCGEPLTPDSSCRPGSPTLSP 257
Query: 543 NHERAAKRQRGS 578
HERAAKRQRGS
Sbjct: 258 KHERAAKRQRGS 269
>sptr|Q84QT0|Q84QT0 Putative transfactor.
Length = 307
Score = 199 bits (506), Expect = 2e-50
Identities = 109/192 (56%), Positives = 125/192 (65%)
Frame = +3
Query: 3 LTIYHVKSHLQKYRLAKYIPDASTDGNKTDNKDPGDLLAGLEGSSGLQISEALKLQMEVQ 182
LTIYHVKSHLQKYRLAKYIPD S D NK ++KDPG+LL+ LEGSSG+QISEALKLQMEVQ
Sbjct: 85 LTIYHVKSHLQKYRLAKYIPDPSADDNKDEDKDPGNLLSALEGSSGMQISEALKLQMEVQ 144
Query: 183 KXXXXXXXXXXXXXXXXXAQGKYLQKIIEEQQRLTGVKSETPAGGASVTVSSDQFPDSER 362
K AQGKYLQKIIEEQQR+ G GAS SS+Q PDSE+
Sbjct: 145 KRLHEQLEVQRQLQLRIEAQGKYLQKIIEEQQRVIG-------AGASRATSSEQLPDSEK 197
Query: 363 XXXXXXXXXXXXXXQVGASNRDTGDRTEATKSTCHGDSLSRNEPLTPDSNCQNGSPVASP 542
Q ++++ + E TKS H D+L EPLTPDS+C+ GSP SP
Sbjct: 198 TNPPTPVPISESPVQGAPHSKNSQSQVEPTKSPSHDDALPCGEPLTPDSSCRPGSPTLSP 257
Query: 543 NHERAAKRQRGS 578
HERAAKRQRGS
Sbjct: 258 KHERAAKRQRGS 269
>sptr|Q9SJW0|Q9SJW0 Transfactor-like protein.
Length = 286
Score = 154 bits (388), Expect = 1e-36
Identities = 94/191 (49%), Positives = 112/191 (58%), Gaps = 1/191 (0%)
Frame = +3
Query: 3 LTIYHVKSHLQKYRLAKYIPDASTDGNKTDNKDPGDLLAGLEGSSGLQISEALKLQMEVQ 182
LTIYHVKSHLQKYRLAKY+PD+S++G KTD K+ GD+L+GL+GSSG+QI+EALKLQMEVQ
Sbjct: 55 LTIYHVKSHLQKYRLAKYLPDSSSEGKKTDKKESGDMLSGLDGSSGMQITEALKLQMEVQ 114
Query: 183 KXXXXXXXXXXXXXXXXXAQGKYLQKIIEEQQRLTGVKSETPAGGASVTVSSDQFPDSER 362
K AQGKYL+KIIEEQQRL+GV E A + + P SE
Sbjct: 115 KRLHEQLEVQRQLQLRIEAQGKYLKKIIEEQQRLSGVLGEPSAPVTGDSDPATPAPTSES 174
Query: 363 XXXXXXXXXXXXXXQVGASNRDTGDRTEATKSTCHGDSLSR-NEPLTPDSNCQNGSPVAS 539
S +D G KS +SLS EPLTPDS C GSP S
Sbjct: 175 PLQ-------------DKSGKDCG----PDKSLSVDESLSSYREPLTPDSGCNIGSPDES 217
Query: 540 PNHERAAKRQR 572
ER +K+ R
Sbjct: 218 TGEERLSKKPR 228
>sptr|Q7XPK2|Q7XPK2 OSJNBa0087O24.8 protein.
Length = 419
Score = 78.6 bits (192), Expect = 5e-14
Identities = 39/91 (42%), Positives = 60/91 (65%)
Frame = +3
Query: 3 LTIYHVKSHLQKYRLAKYIPDASTDGNKTDNKDPGDLLAGLEGSSGLQISEALKLQMEVQ 182
LTIYH+KSHLQKYR+AKY+P AS++G + + + G+ + L+ +G+QI+EAL++Q++VQ
Sbjct: 276 LTIYHIKSHLQKYRIAKYMP-ASSEGKQLEKRATGNDMQNLDPKTGMQITEALRVQLDVQ 334
Query: 183 KXXXXXXXXXXXXXXXXXAQGKYLQKIIEEQ 275
+ QGK LQK+ E+Q
Sbjct: 335 RRLHEQLEIQRNLQLRIEEQGKRLQKMFEDQ 365
>sptr|Q949U2|Q949U2 Hypothetical protein.
Length = 449
Score = 76.6 bits (187), Expect = 2e-13
Identities = 49/124 (39%), Positives = 69/124 (55%), Gaps = 5/124 (4%)
Frame = +3
Query: 3 LTIYHVKSHLQKYRLAKYIPDASTDGNKTDNKDPGDLL-----AGLEGSSGLQISEALKL 167
LTIYHVKSHLQKYRLAKY+P+ + +TDN + L A + +Q++EAL++
Sbjct: 281 LTIYHVKSHLQKYRLAKYMPEKKEE-KRTDNSEEKKLALSKSEADEKKKGAIQLTEALRM 339
Query: 168 QMEVQKXXXXXXXXXXXXXXXXXAQGKYLQKIIEEQQRLTGVKSETPAGGASVTVSSDQF 347
QMEVQK KYL+K++EE QR TG + + + ++ S D
Sbjct: 340 QMEVQKQLHEQLEVQRVLQLRIEEHAKYLEKMLEE-QRKTG-RWISSSSQTVLSPSDDSI 397
Query: 348 PDSE 359
PDS+
Sbjct: 398 PDSQ 401
>sptr|Q8LFN4|Q8LFN4 Transfactor-like protein.
Length = 443
Score = 76.6 bits (187), Expect = 2e-13
Identities = 49/124 (39%), Positives = 69/124 (55%), Gaps = 5/124 (4%)
Frame = +3
Query: 3 LTIYHVKSHLQKYRLAKYIPDASTDGNKTDNKDPGDLL-----AGLEGSSGLQISEALKL 167
LTIYHVKSHLQKYRLAKY+P+ + +TDN + L A + +Q++EAL++
Sbjct: 275 LTIYHVKSHLQKYRLAKYMPEKKEE-KRTDNSEEKKLALSKSEADEKKKGAIQLTEALRM 333
Query: 168 QMEVQKXXXXXXXXXXXXXXXXXAQGKYLQKIIEEQQRLTGVKSETPAGGASVTVSSDQF 347
QMEVQK KYL+K++EE QR TG + + + ++ S D
Sbjct: 334 QMEVQKQLHEQQEVQRVLQLRIEEHAKYLEKMLEE-QRKTG-RWISSSSQTVLSPSDDSI 391
Query: 348 PDSE 359
PDS+
Sbjct: 392 PDSQ 395
>sptr|Q9LSH8|Q9LSH8 Transfactor-like protein.
Length = 554
Score = 76.6 bits (187), Expect = 2e-13
Identities = 49/124 (39%), Positives = 69/124 (55%), Gaps = 5/124 (4%)
Frame = +3
Query: 3 LTIYHVKSHLQKYRLAKYIPDASTDGNKTDNKDPGDLL-----AGLEGSSGLQISEALKL 167
LTIYHVKSHLQKYRLAKY+P+ + +TDN + L A + +Q++EAL++
Sbjct: 281 LTIYHVKSHLQKYRLAKYMPEKKEE-KRTDNSEEKKLALSKSEADEKKKGAIQLTEALRM 339
Query: 168 QMEVQKXXXXXXXXXXXXXXXXXAQGKYLQKIIEEQQRLTGVKSETPAGGASVTVSSDQF 347
QMEVQK KYL+K++EE QR TG + + + ++ S D
Sbjct: 340 QMEVQKQLHEQLEVQRVLQLRIEEHAKYLEKMLEE-QRKTG-RWISSSSQTVLSPSDDSI 397
Query: 348 PDSE 359
PDS+
Sbjct: 398 PDSQ 401
>sptr|Q9ZQT9|Q9ZQT9 WERBP-1 protein.
Length = 291
Score = 76.3 bits (186), Expect = 3e-13
Identities = 42/91 (46%), Positives = 56/91 (61%)
Frame = +3
Query: 3 LTIYHVKSHLQKYRLAKYIPDASTDGNKTDNKDPGDLLAGLEGSSGLQISEALKLQMEVQ 182
LTIYHVKSHLQKYR A+Y P+A ++ GDL A L+ +G++I+EAL+LQMEVQ
Sbjct: 112 LTIYHVKSHLQKYRTARYKPEALEGSSEKKESSIGDLSA-LDLKTGIEITEALRLQMEVQ 170
Query: 183 KXXXXXXXXXXXXXXXXXAQGKYLQKIIEEQ 275
K QG+YLQ++ E+Q
Sbjct: 171 KQLHEQLEIQRNLQLRIEEQGRYLQEMFEKQ 201
>sptr|Q94CL7|Q94CL7 Phosphate starvation response regulator 1
(Hypothetical protein).
Length = 409
Score = 75.9 bits (185), Expect = 3e-13
Identities = 45/114 (39%), Positives = 60/114 (52%)
Frame = +3
Query: 3 LTIYHVKSHLQKYRLAKYIPDASTDGNKTDNKDPGDLLAGLEGSSGLQISEALKLQMEVQ 182
LTIYHVKSHLQKYR A+Y P+ S G+ P + + L+ G+ I+EAL+LQMEVQ
Sbjct: 265 LTIYHVKSHLQKYRTARYRPEPSETGSPERKLTPLEHITSLDLKGGIGITEALRLQMEVQ 324
Query: 183 KXXXXXXXXXXXXXXXXXAQGKYLQKIIEEQQRLTGVKSETPAGGASVTVSSDQ 344
K QGKYLQ + E+Q +G+ T + S S +
Sbjct: 325 KQLHEQLEIQRNLQLRIEEQGKYLQMMFEKQN--SGLTKGTASTSDSAAKSEQE 376
>sptr|Q9M842|Q9M842 Transfactor, putative.
Length = 438
Score = 74.7 bits (182), Expect = 8e-13
Identities = 50/154 (32%), Positives = 74/154 (48%), Gaps = 1/154 (0%)
Frame = +3
Query: 3 LTIYHVKSHLQKYRLAKYIPDASTDGNKTDNKDPGDLLAGLEGSSGLQISEALKLQMEVQ 182
LT+YHVKSHLQKYR A+Y P+ S D K N + + L+ + ++I+EAL+LQM+VQ
Sbjct: 278 LTVYHVKSHLQKYRTARYKPELSKDTVK--NLKTIEDIKSLDLKTSIEITEALRLQMKVQ 335
Query: 183 KXXXXXXXXXXXXXXXXXAQGKYLQKIIEEQQRL-TGVKSETPAGGASVTVSSDQFPDSE 359
K QG+YLQ +IE+QQ++ K T + S P+
Sbjct: 336 KQLHEQLEIQRSLQLQIEEQGRYLQMMIEKQQKMQENKKDSTSSSSMPEADPSAPSPNLS 395
Query: 360 RXXXXXXXXXXXXXXQVGASNRDTGDRTEATKST 461
+ Q + T D++E+T T
Sbjct: 396 QPFLHKATNSEPSITQKLQNGSSTMDQSESTSGT 429
>sptr|Q8GWU5|Q8GWU5 Putative transfactor (At3g04445).
Length = 402
Score = 73.9 bits (180), Expect = 1e-12
Identities = 49/156 (31%), Positives = 74/156 (47%), Gaps = 3/156 (1%)
Frame = +3
Query: 3 LTIYHVKSHLQKYRLAKYIPDASTDGNK--TDNKDPGDLLAGLEGSSGLQISEALKLQME 176
LT+YHVKSHLQKYR A+Y P+ S D + N + + L+ + ++I+EAL+LQM+
Sbjct: 238 LTVYHVKSHLQKYRTARYKPELSKDTEEPLVKNLKTIEDIKSLDLKTSIEITEALRLQMK 297
Query: 177 VQKXXXXXXXXXXXXXXXXXAQGKYLQKIIEEQQRL-TGVKSETPAGGASVTVSSDQFPD 353
VQK QG+YLQ +IE+QQ++ K T + S P+
Sbjct: 298 VQKQLHEQLEIQRSLQLQIEEQGRYLQMMIEKQQKMQENKKDSTSSSSMPEADPSAPSPN 357
Query: 354 SERXXXXXXXXXXXXXXQVGASNRDTGDRTEATKST 461
+ Q + T D++E+T T
Sbjct: 358 LSQPFLHKATNSEPSITQKLQNGSSTMDQSESTSGT 393
>sptrnew|BAD10540|BAD10540 Putative transfactor.
Length = 467
Score = 73.6 bits (179), Expect = 2e-12
Identities = 41/95 (43%), Positives = 55/95 (57%), Gaps = 2/95 (2%)
Frame = +3
Query: 3 LTIYHVKSHLQKYRLAKYIPDASTDGNKTDNKDPGDLLAGLEGS--SGLQISEALKLQME 176
LTIYH+KSHLQKYRLAKY+P+ D K + K + G + + Q++EAL++QME
Sbjct: 305 LTIYHIKSHLQKYRLAKYLPETKED-KKQEEKKTKSVANGNDHAKKKSAQMAEALRMQME 363
Query: 177 VQKXXXXXXXXXXXXXXXXXAQGKYLQKIIEEQQR 281
VQK +YLQKI+EEQQ+
Sbjct: 364 VQKQLHEQLEVQRQLQLRIEEHARYLQKILEEQQK 398
>sptr|Q94AL7|Q94AL7 Hypothetical protein (Fragment).
Length = 385
Score = 72.0 bits (175), Expect = 5e-12
Identities = 40/103 (38%), Positives = 58/103 (56%), Gaps = 2/103 (1%)
Frame = +3
Query: 3 LTIYHVKSHLQKYRLAKYIPDASTDGNKTDNKDPGDL--LAGLEGSSGLQISEALKLQME 176
LTIYHVKSHLQKYR A+Y P+ S + K + + L+ + ++I++AL+LQME
Sbjct: 243 LTIYHVKSHLQKYRTARYKPETSEVTGEPQEKKMTSIEDIKSLDMKTSVEITQALRLQME 302
Query: 177 VQKXXXXXXXXXXXXXXXXXAQGKYLQKIIEEQQRLTGVKSET 305
VQK QG+YLQ + E+QQ++ KS +
Sbjct: 303 VQKRLHEQLEIQRSLQLQIEKQGRYLQMMFEKQQKIQDNKSSS 345
>sptr|Q8GUN5|Q8GUN5 Hypothetical protein.
Length = 413
Score = 72.0 bits (175), Expect = 5e-12
Identities = 40/103 (38%), Positives = 58/103 (56%), Gaps = 2/103 (1%)
Frame = +3
Query: 3 LTIYHVKSHLQKYRLAKYIPDASTDGNKTDNKDPGDL--LAGLEGSSGLQISEALKLQME 176
LTIYHVKSHLQKYR A+Y P+ S + K + + L+ + ++I++AL+LQME
Sbjct: 271 LTIYHVKSHLQKYRTARYKPETSEVTGEPQEKKMTSIEDIKSLDMKTSVEITQALRLQME 330
Query: 177 VQKXXXXXXXXXXXXXXXXXAQGKYLQKIIEEQQRLTGVKSET 305
VQK QG+YLQ + E+QQ++ KS +
Sbjct: 331 VQKRLHEQLEIQRSLQLQIEKQGRYLQMMFEKQQKIQDNKSSS 373
>sptr|Q8LEY3|Q8LEY3 Transfactor, putative.
Length = 413
Score = 72.0 bits (175), Expect = 5e-12
Identities = 40/103 (38%), Positives = 58/103 (56%), Gaps = 2/103 (1%)
Frame = +3
Query: 3 LTIYHVKSHLQKYRLAKYIPDASTDGNKTDNKDPGDL--LAGLEGSSGLQISEALKLQME 176
LTIYHVKSHLQKYR A+Y P+ S + K + + L+ + ++I++AL+LQME
Sbjct: 271 LTIYHVKSHLQKYRTARYKPETSEVTGEPQEKKMTSIEDIKSLDMKTSVEITQALRLQME 330
Query: 177 VQKXXXXXXXXXXXXXXXXXAQGKYLQKIIEEQQRLTGVKSET 305
VQK QG+YLQ + E+QQ++ KS +
Sbjct: 331 VQKRLHEQLEIQRSLQLQIEKQGRYLQMMFEKQQKIQDNKSSS 373
>sptr|Q9M620|Q9M620 CDPK substrate protein 1.
Length = 470
Score = 70.1 bits (170), Expect = 2e-11
Identities = 42/120 (35%), Positives = 61/120 (50%)
Frame = +3
Query: 3 LTIYHVKSHLQKYRLAKYIPDASTDGNKTDNKDPGDLLAGLEGSSGLQISEALKLQMEVQ 182
LTIYHVKSHLQKYR A+ P+ S++GN D ++ ++ + + I+EAL++QMEVQ
Sbjct: 296 LTIYHVKSHLQKYRTARVRPE-SSEGNSERRASSVDPVSSVDLKTSVTITEALRMQMEVQ 354
Query: 183 KXXXXXXXXXXXXXXXXXAQGKYLQKIIEEQQRLTGVKSETPAGGASVTVSSDQFPDSER 362
K QGKYL +++E Q ++ K A S P+ R
Sbjct: 355 KQLHEQLEIQRKLQLQIEEQGKYLLQMLENQNKVEKEKLNPDGSSAHNDKSEGSQPEPSR 414
>sptr|Q9FG27|Q9FG27 Similarity to transfactor.
Length = 374
Score = 68.9 bits (167), Expect = 4e-11
Identities = 37/93 (39%), Positives = 57/93 (61%)
Frame = +3
Query: 3 LTIYHVKSHLQKYRLAKYIPDASTDGNKTDNKDPGDLLAGLEGSSGLQISEALKLQMEVQ 182
LTI+HVKSHLQKYR+AKY+P++ K + + L+ L+ +G+QI EAL+LQ++VQ
Sbjct: 231 LTIFHVKSHLQKYRIAKYMPESQE--GKFEKRACAKELSQLDTRTGVQIKEALQLQLDVQ 288
Query: 183 KXXXXXXXXXXXXXXXXXAQGKYLQKIIEEQQR 281
+ QGK L+ ++E+QQ+
Sbjct: 289 RHLHEQLEIQRNLQLRIEEQGKQLKMMMEQQQK 321
>sptr|Q9M0H0|Q9M0H0 Hypothetical protein.
Length = 422
Score = 68.9 bits (167), Expect = 4e-11
Identities = 46/127 (36%), Positives = 61/127 (48%), Gaps = 13/127 (10%)
Frame = +3
Query: 3 LTIYHVKSHLQKYRLAKYIPDAS-------------TDGNKTDNKDPGDLLAGLEGSSGL 143
LTIYHVKSHLQKYR A+Y P+ S T G+ P + + L+ G+
Sbjct: 265 LTIYHVKSHLQKYRTARYRPEPSETEFNVKTKVSLITTGSPERKLTPLEHITSLDLKGGI 324
Query: 144 QISEALKLQMEVQKXXXXXXXXXXXXXXXXXAQGKYLQKIIEEQQRLTGVKSETPAGGAS 323
I+EAL+LQMEVQK QGKYLQ + E+Q +G+ T + S
Sbjct: 325 GITEALRLQMEVQKQLHEQLEIQRNLQLRIEEQGKYLQMMFEKQN--SGLTKGTASTSDS 382
Query: 324 VTVSSDQ 344
S +
Sbjct: 383 AAKSEQE 389
>sptr|Q9LRN5|Q9LRN5 Similarity to transfactor.
Length = 307
Score = 66.6 bits (161), Expect = 2e-10
Identities = 41/111 (36%), Positives = 61/111 (54%), Gaps = 7/111 (6%)
Frame = +3
Query: 3 LTIYHVKSHLQKYRLAKYIPDASTDGNKT-DNKDPGD------LLAGLEGSSGLQISEAL 161
LT+YH+KSHLQK+RL + ST+ +K +++D G +A E + G Q++EAL
Sbjct: 97 LTLYHLKSHLQKFRLGRQAGKESTENSKDGESQDTGSSSTSSMRMAQQEQNEGYQVTEAL 156
Query: 162 KLQMEVQKXXXXXXXXXXXXXXXXXAQGKYLQKIIEEQQRLTGVKSETPAG 314
+ QMEVQ+ AQGKYLQ I+E+ + ++ T AG
Sbjct: 157 RAQMEVQRRLHDQLEVQRRLQLRIEAQGKYLQSILEKACKAFDEQAATFAG 207
>sptr|Q84WH5|Q84WH5 Transfactor-like protein.
Length = 402
Score = 65.5 bits (158), Expect = 5e-10
Identities = 49/140 (35%), Positives = 67/140 (47%), Gaps = 22/140 (15%)
Frame = +3
Query: 3 LTIYHVKSHLQKYRLAKYI-PDASTDGNKT-------------DNKDPGDLLAGLEGSSG 140
LT+YH+KSHLQKYRL+K + A++ NKT D L G + S
Sbjct: 85 LTLYHLKSHLQKYRLSKNLNGQANSSLNKTSVMTMVEENPPEVDESHSESLSIGPQPSMN 144
Query: 141 LQISEALKLQMEVQKXXXXXXXXXXXXXXXXXAQGKYLQKIIEEQQRLTGVKSETPAG-- 314
L IS+AL++Q+EVQ+ AQGKYLQ I+E+ Q G ++ AG
Sbjct: 145 LPISDALQMQIEVQRRLHEQLEVQRHLQLRIEAQGKYLQSILEKAQETLGRQNLGAAGIE 204
Query: 315 ------GASVTVSSDQFPDS 356
V+ S +PDS
Sbjct: 205 ATKAQLSELVSKVSADYPDS 224
>sptr|Q94B34|Q94B34 Transfactor-like protein.
Length = 402
Score = 65.5 bits (158), Expect = 5e-10
Identities = 49/140 (35%), Positives = 67/140 (47%), Gaps = 22/140 (15%)
Frame = +3
Query: 3 LTIYHVKSHLQKYRLAKYI-PDASTDGNKT-------------DNKDPGDLLAGLEGSSG 140
LT+YH+KSHLQKYRL+K + A++ NKT D L G + S
Sbjct: 85 LTLYHLKSHLQKYRLSKNLNGQANSSLNKTSVMTMVEENPPEVDESHSESLSIGPQPSMN 144
Query: 141 LQISEALKLQMEVQKXXXXXXXXXXXXXXXXXAQGKYLQKIIEEQQRLTGVKSETPAG-- 314
L IS+AL++Q+EVQ+ AQGKYLQ I+E+ Q G ++ AG
Sbjct: 145 LPISDALQMQIEVQRRLHEQLEVQRHLQLRIEAQGKYLQSILEKAQETLGRQNLGAAGIE 204
Query: 315 ------GASVTVSSDQFPDS 356
V+ S +PDS
Sbjct: 205 ATKAQLSELVSKVSADYPDS 224
>sptr|Q9FK47|Q9FK47 Transfactor-like protein.
Length = 402
Score = 65.5 bits (158), Expect = 5e-10
Identities = 49/140 (35%), Positives = 67/140 (47%), Gaps = 22/140 (15%)
Frame = +3
Query: 3 LTIYHVKSHLQKYRLAKYI-PDASTDGNKT-------------DNKDPGDLLAGLEGSSG 140
LT+YH+KSHLQKYRL+K + A++ NKT D L G + S
Sbjct: 85 LTLYHLKSHLQKYRLSKNLNGQANSSLNKTSVMTMVEENPPEVDESHSESLSIGPQPSMN 144
Query: 141 LQISEALKLQMEVQKXXXXXXXXXXXXXXXXXAQGKYLQKIIEEQQRLTGVKSETPAG-- 314
L IS+AL++Q+EVQ+ AQGKYLQ I+E+ Q G ++ AG
Sbjct: 145 LPISDALQMQIEVQRRLHEQLEVQRHLQLRIEAQGKYLQSILEKAQETLGRQNLGAAGIE 204
Query: 315 ------GASVTVSSDQFPDS 356
V+ S +PDS
Sbjct: 205 ATKAQLSELVSKVSADYPDS 224
>sptr|Q94A57|Q94A57 AT3g24120/MUJ8_3 (Transfactor, putative).
Length = 295
Score = 65.1 bits (157), Expect = 6e-10
Identities = 41/115 (35%), Positives = 61/115 (53%), Gaps = 11/115 (9%)
Frame = +3
Query: 3 LTIYHVKSHLQKYRLAKYIPDASTDGNKT-----DNKDPGD------LLAGLEGSSGLQI 149
LT+YH+KSHLQK+RL + ST+ +K +++D G +A E + G Q+
Sbjct: 81 LTLYHLKSHLQKFRLGRQAGKESTENSKDASCVGESQDTGSSSTSSMRMAQQEQNEGYQV 140
Query: 150 SEALKLQMEVQKXXXXXXXXXXXXXXXXXAQGKYLQKIIEEQQRLTGVKSETPAG 314
+EAL+ QMEVQ+ AQGKYLQ I+E+ + ++ T AG
Sbjct: 141 TEALRAQMEVQRRLHDQLEVQRRLQLRIEAQGKYLQSILEKACKAFDEQAATFAG 195
>sptr|Q9SEZ2|Q9SEZ2 Transfactor, putative, 28697-27224.
Length = 332
Score = 63.9 bits (154), Expect = 1e-09
Identities = 42/116 (36%), Positives = 59/116 (50%), Gaps = 17/116 (14%)
Frame = +3
Query: 3 LTIYHVKSHLQKYRLAKYIP--------DASTDGNKTDNKDPGDLLAGLE--------GS 134
LT+YH+KSHLQKYRL K + ++++ + ++K+ L G
Sbjct: 70 LTLYHLKSHLQKYRLGKSMKFDDNKLEVSSASENQEVESKNDSRDLRGCSVTEENSNPAK 129
Query: 135 SGLQISEALKLQMEVQKXXXXXXXXXXXXXXXXXAQGKYLQKII-EEQQRLTGVKS 299
GLQI+EAL++QMEVQK AQGKYLQ ++ + QQ L G S
Sbjct: 130 EGLQITEALQMQMEVQKKLHEQIEVQRHLQVKIEAQGKYLQSVLMKAQQTLAGYSS 185
>sptr|Q8GXC2|Q8GXC2 Hypothetical protein.
Length = 397
Score = 63.2 bits (152), Expect = 2e-09
Identities = 41/118 (34%), Positives = 58/118 (49%), Gaps = 7/118 (5%)
Frame = +3
Query: 3 LTIYHVKSHLQKYRLAKYIPDASTDGNKTDNKDPGDLLAGLEGSSGLQISEALKLQMEVQ 182
LTI+HVKSHLQKYR AKYIP ++G+ P + + + G+ I+E L++QME Q
Sbjct: 271 LTIFHVKSHLQKYRTAKYIP-VPSEGSPEARLTPLEQITSDDTKRGIDITETLRIQMEHQ 329
Query: 183 KXXXXXXXXXXXXXXXXXAQGKYLQKIIEEQQRLTG-------VKSETPAGGASVTVS 335
K QGK L +IE+Q G ++TP G+ + S
Sbjct: 330 KKLHEQLESLRTMQLRIEEQGKALLMMIEKQNMGFGGPEQGEKTSAKTPENGSEESES 387
>sptrnew|BAC84294|BAC84294 Putative CDPK substrate protein 1.
Length = 426
Score = 62.8 bits (151), Expect = 3e-09
Identities = 40/118 (33%), Positives = 57/118 (48%)
Frame = +3
Query: 3 LTIYHVKSHLQKYRLAKYIPDASTDGNKTDNKDPGDLLAGLEGSSGLQISEALKLQMEVQ 182
LTIYHVKSHLQKYR A+Y P+ S ++ D+ + ++EAL+LQ+E+Q
Sbjct: 286 LTIYHVKSHLQKYRTARYRPELSEGSSEKKAASKEDIPSIDLKGGNFDLTEALRLQLELQ 345
Query: 183 KXXXXXXXXXXXXXXXXXAQGKYLQKIIEEQQRLTGVKSETPAGGASVTVSSDQFPDS 356
K QGK LQ ++E+Q K+ + A T S P+S
Sbjct: 346 KRLHEQLEIQRSLQLRIEEQGKCLQMMLEQQCIPGTDKAVDASTSAEGTKPSSDLPES 403
>sptr|Q8LAJ7|Q8LAJ7 Transfactor, putative.
Length = 292
Score = 61.6 bits (148), Expect = 7e-09
Identities = 38/101 (37%), Positives = 53/101 (52%), Gaps = 11/101 (10%)
Frame = +3
Query: 3 LTIYHVKSHLQKYRLAKYIPDASTDGNK-----TDNKDPGD------LLAGLEGSSGLQI 149
LT+YH+KSHLQK+RL + S D +K +++D G LA E + Q+
Sbjct: 77 LTLYHLKSHLQKFRLGRQSCKESIDNSKDVSCVAESQDTGSSSTSSLRLAAQEQNESYQV 136
Query: 150 SEALKLQMEVQKXXXXXXXXXXXXXXXXXAQGKYLQKIIEE 272
+EAL+ QMEVQ+ AQGKYLQ I+E+
Sbjct: 137 TEALRAQMEVQRRLHEQLEVQRRLQLRIEAQGKYLQSILEK 177
>sptr|Q9C5Y6|Q9C5Y6 MYR1.
Length = 396
Score = 61.2 bits (147), Expect = 9e-09
Identities = 49/140 (35%), Positives = 67/140 (47%), Gaps = 22/140 (15%)
Frame = +3
Query: 3 LTIYHVKSHLQKYRLAKYI-PDASTDGNKT-------------DNKDPGDLLAGLEGSSG 140
LT+YH+KSHLQKYRL+K + A++ NKT D L G + S
Sbjct: 85 LTLYHLKSHLQKYRLSKNLNGQANSSLNKTSVMTMVEENPPEVDESHSESLSIGPQPSMN 144
Query: 141 LQISEALKLQMEVQKXXXXXXXXXXXXXXXXXAQGKYLQKIIEEQQRLTGVKSETPAG-- 314
L IS+AL++Q+EVQ+ AQGKYLQ I+E+ Q G ++ AG
Sbjct: 145 LPISDALQMQIEVQR------RLHEQLELRIEAQGKYLQSILEKAQETLGRQNLGAAGIE 198
Query: 315 ------GASVTVSSDQFPDS 356
V+ S +PDS
Sbjct: 199 ATKAQLSELVSKVSADYPDS 218
>sptr|Q9SQQ9|Q9SQQ9 Transfactor-like.
Length = 394
Score = 59.3 bits (142), Expect = 3e-08
Identities = 46/140 (32%), Positives = 67/140 (47%), Gaps = 22/140 (15%)
Frame = +3
Query: 3 LTIYHVKSHLQKYRLAKYI-PDASTDGNK-------------TDNKDPGDLLAGLEGSSG 140
LT+YH+KSHLQKYRL+K + A+ NK D +L G + +
Sbjct: 85 LTLYHLKSHLQKYRLSKNLNGQANNSFNKIGIMTMMEEKTPDADEIQSENLSIGPQPNKN 144
Query: 141 LQISEALKLQMEVQKXXXXXXXXXXXXXXXXXAQGKYLQKIIEEQQRLTGVKSETPAG-- 314
I EAL++Q+EVQ+ AQGKYLQ ++E+ Q G ++ AG
Sbjct: 145 SPIGEALQMQIEVQRRLHEQLEVQRHLQLRIEAQGKYLQSVLEKAQETLGRQNLGAAGIE 204
Query: 315 GASVTVS------SDQFPDS 356
A V +S S ++P+S
Sbjct: 205 AAKVQLSELVSKVSAEYPNS 224
>sptr|Q7X8E9|Q7X8E9 Putative phosphate starvation response regulator
(Putative calcium-dependent protein kinase substrate
protein).
Length = 282
Score = 57.4 bits (137), Expect = 1e-07
Identities = 37/116 (31%), Positives = 53/116 (45%), Gaps = 13/116 (11%)
Frame = +3
Query: 3 LTIYHVKSHLQKYRLAKYIPDASTDGNKTDN------------KDPGDLLAGLEGSSG-L 143
LT+YH+KSHLQKYRL + + +N P + ++G G
Sbjct: 66 LTLYHLKSHLQKYRLGQQTKKQNAAEQNRENIGESFRQFSLHSSGPSITSSSMDGMQGEA 125
Query: 144 QISEALKLQMEVQKXXXXXXXXXXXXXXXXXAQGKYLQKIIEEQQRLTGVKSETPA 311
ISEAL+ Q+EVQK AQGKYLQ I+++ Q+ +P+
Sbjct: 126 PISEALRCQIEVQKRLHEQLEVQQKLQMRIEAQGKYLQAILDKAQKSLSTDMNSPS 181
Database: /db/uniprot/tmp/swall
Posted date: Mar 5, 2004 7:45 PM
Number of letters in database: 442,889,342
Number of sequences in database: 1,395,590
Lambda K H
0.318 0.135 0.401
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 472,129,954
Number of Sequences: 1395590
Number of extensions: 9336324
Number of successful extensions: 30368
Number of sequences better than 10.0: 30
Number of HSP's better than 10.0 without gapping: 29370
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 30281
length of database: 442,889,342
effective HSP length: 116
effective length of database: 281,000,902
effective search space used: 21637069454
frameshift window, decay const: 50, 0.1
T: 12
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)

