BLASTX 2.2.6 [Apr-09-2003]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= 4580138.2.1
(1484 letters)
Database: /db/uniprot/tmp/swall
1,395,590 sequences; 442,889,342 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
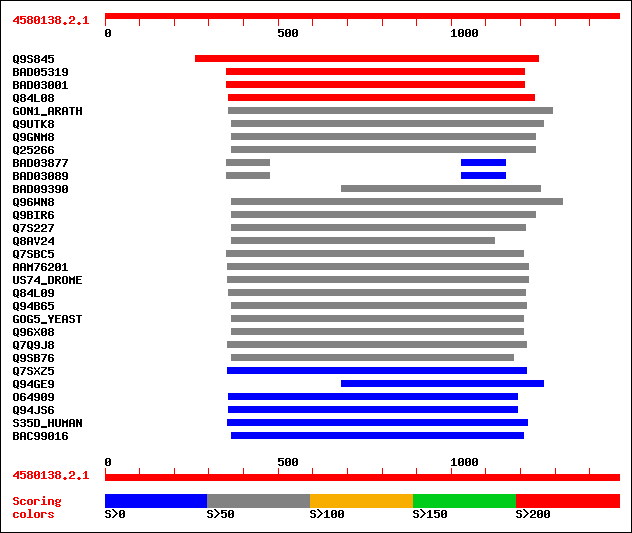
sptr|Q9S845|Q9S845 T23E18.26 (GONST3 golgi nucleotide sugar tran... 457 e-127
sptrnew|BAD05319|BAD05319 Putative integral membrane protein. 295 1e-78
sptrnew|BAD03001|BAD03001 Putative integral membrane protein. 295 1e-78
sptr|Q84L08|Q84L08 GONST4 golgi nucleotide sugar transporter (Fr... 267 3e-70
sw|Q941R4|GON1_ARATH GDP-mannose transporter. 74 5e-12
sptr|Q9UTK8|Q9UTK8 Putative vanadate resistance protein. 65 3e-09
sptr|Q9GNM8|Q9GNM8 LPG2 protein. 64 8e-09
sptr|Q25266|Q25266 LPG2. 62 2e-08
sptrnew|BAD03877|BAD03877 Hypothetical protein OJ1118_F01.31. 62 3e-08
sptrnew|BAD03089|BAD03089 Hypothetical protein OJ1484_G09.115. 62 3e-08
sptrnew|BAD09390|BAD09390 Putative GDP-Mannose transporter. 59 2e-07
sptr|Q96WN8|Q96WN8 Golgi GDP-mannose transporter. 59 3e-07
sptr|Q9BIR6|Q9BIR6 Lipophosphoglycan biosynthetic protein. 58 3e-07
sptr|Q7S227|Q7S227 Hypothetical protein. 56 2e-06
sptr|Q8AV24|Q8AV24 UGNT. 55 2e-06
sptr|Q7SBC5|Q7SBC5 Hypothetical protein. 55 2e-06
sptrnew|AAM76201|AAM76201 RH08077p. 55 3e-06
sw|Q95YI5|US74_DROME UDP-sugar transporter UST74c (Fringe connec... 55 3e-06
sptr|Q84L09|Q84L09 GONST2 golgi nucleotide sugar transporter (Fr... 55 3e-06
sptr|Q94B65|Q94B65 Hypothetical protein. 55 4e-06
sw|P40107|GOG5_YEAST GDP-mannose transporter. 54 8e-06
sptr|Q96X08|Q96X08 GDP-mannose transporter. 53 1e-05
sptr|Q7Q9J8|Q7Q9J8 AgCP15152 (Fragment). 52 3e-05
sptr|Q9SB76|Q9SB76 Hypothetical protein. 51 5e-05
sptr|Q7SXZ5|Q7SXZ5 Hypothetical protein. 48 5e-04
sptr|Q94GE9|Q94GE9 Putative lipophosphoglycan biosynthetic. 47 6e-04
sptr|O64909|O64909 Glucose-6-phosphate/phosphate-translocator pr... 47 8e-04
sptr|Q94JS6|Q94JS6 Glucose-6-phosphate/phosphate translocator. 46 0.002
sw|Q9NTN3|S35D_HUMAN UDP-glucuronic acid/UDP-N-acetylgalactosami... 46 0.002
sptrnew|BAC99016|BAC99016 Nucleotide sugar transporter UGTrel8. 45 0.002
>sptr|Q9S845|Q9S845 T23E18.26 (GONST3 golgi nucleotide sugar
transporter).
Length = 372
Score = 457 bits (1177), Expect = e-127
Identities = 229/332 (68%), Positives = 268/332 (80%), Gaps = 1/332 (0%)
Frame = -1
Query: 1253 NTLMQQASIYGVAAGYCLSASLLSIINKWAIMKFPYPGALTALQYFTSVAGVLLCGQLKL 1074
+ ++QAS+YGVAAGYCLSASLLSIINKWAIMKFPYPGALTA+QYFTS AGVLLC Q+KL
Sbjct: 27 SVFLRQASVYGVAAGYCLSASLLSIINKWAIMKFPYPGALTAMQYFTSAAGVLLCAQMKL 86
Query: 1073 IEHDGLNLRTMWKFLPAAVMFYISIFTNSELLLHADVDTFIVFRSAVPIFVAIGETLYLH 894
IEHD LNL TMW+FLPAA++FY+S+FTNSELLLHA+VDTFIVFRSAVPIFVAIGETL+LH
Sbjct: 87 IEHDSLNLLTMWRFLPAAMIFYLSLFTNSELLLHANVDTFIVFRSAVPIFVAIGETLFLH 146
Query: 893 QPWPSLKTWLSLSTILGGSVIYVFTDNQFTVTAYTWAVAYLASMSIDFVYIKHVVMTIGL 714
QPWPS+KTW SL+TI GGS++YVFTD QFT+ AY+WA+AYL SM+IDFVYIKHVVMTIGL
Sbjct: 147 QPWPSVKTWGSLATIFGGSLLYVFTDYQFTIAAYSWALAYLVSMTIDFVYIKHVVMTIGL 206
Query: 713 NTWGLVLYNNXXXXXXXXXXXLVMGEFDQMKVDSSKVSNWLSFDVVLPVAXXXXXXXXXX 534
NTWGLVLYNN L+MGE ++K + + ++W S VVLPV
Sbjct: 207 NTWGLVLYNNLEALLLFPLELLIMGELKKIKHEITDETDWYSLQVVLPVGLSCLFGLAIS 266
Query: 533 XXXXXCRRAISATGFTVLGIVNKLLTVVINLLIWDKHASFVGTIGLLICMSGGVLYQQST 354
CRRAISATGFTVLGIVNKLLTVVINL++WDKH++FVGT+GLL+CM GGV+YQQST
Sbjct: 267 FFGFSCRRAISATGFTVLGIVNKLLTVVINLMVWDKHSTFVGTLGLLVCMFGGVMYQQST 326
Query: 353 TXX-XXXXXXXXXENDEEQQKLLQMQGVQESS 261
E DEEQ+KLL+MQ +ES+
Sbjct: 327 IKKPNATQEAKPQEQDEEQEKLLEMQENKESN 358
>sptrnew|BAD05319|BAD05319 Putative integral membrane protein.
Length = 342
Score = 295 bits (756), Expect = 1e-78
Identities = 153/288 (53%), Positives = 194/288 (67%), Gaps = 1/288 (0%)
Frame = -1
Query: 1211 GYCLSASLLSIINKWAIMKFPYPGALTALQYFTSVAGVLLCGQLKLIEHDGLNLRTMWKF 1032
GY L +SLL+IINK+AI KF YPG LTALQY TSVAGV G+L L+ HD NL+T KF
Sbjct: 14 GYALCSSLLAIINKYAITKFSYPGLLTALQYLTSVAGVWTLGKLGLLYHDPFNLQTAKKF 73
Query: 1031 LPAAVMFYISIFTNSELLLHADVDTFIVFRSAVPIFVAIGETLYLHQPWPSLKTWLSLST 852
PAA++FY++IFTN+ LL HA+VDTFIVFRS P+ VAI +T + QP PS T++SL T
Sbjct: 74 APAALVFYLAIFTNTHLLKHANVDTFIVFRSLTPLLVAIADTAFRKQPCPSKLTFVSLVT 133
Query: 851 ILGGSVIYVFTDNQFTVTAYTWAVAYLASMSIDFVYIKHVVMTIGLNTWGLVLYNNXXXX 672
ILGG+V YV TD+ F++TAY+WAVAYL +++ + VYIKH+V +GLNTWG VLYNN
Sbjct: 134 ILGGAVGYVMTDSGFSLTAYSWAVAYLVTITTEMVYIKHMVTKLGLNTWGFVLYNNLLSL 193
Query: 671 XXXXXXXLVMGE-FDQMKVDSSKVSNWLSFDVVLPVAXXXXXXXXXXXXXXXCRRAISAT 495
+ GE + S+ +W D + V+ R+AISAT
Sbjct: 194 IIAPVFWFLTGEHLSVFRAIESRGQSWFELDAFVAVSLSCVFGLLISFFGFAARKAISAT 253
Query: 494 GFTVLGIVNKLLTVVINLLIWDKHASFVGTIGLLICMSGGVLYQQSTT 351
FTV G+VNK LTV IN++IWDKHAS G + LL ++GGVLYQQS T
Sbjct: 254 AFTVTGVVNKFLTVAINVMIWDKHASSFGLVCLLFTLAGGVLYQQSVT 301
>sptrnew|BAD03001|BAD03001 Putative integral membrane protein.
Length = 342
Score = 295 bits (756), Expect = 1e-78
Identities = 153/288 (53%), Positives = 194/288 (67%), Gaps = 1/288 (0%)
Frame = -1
Query: 1211 GYCLSASLLSIINKWAIMKFPYPGALTALQYFTSVAGVLLCGQLKLIEHDGLNLRTMWKF 1032
GY L +SLL+IINK+AI KF YPG LTALQY TSVAGV G+L L+ HD NL+T KF
Sbjct: 14 GYALCSSLLAIINKYAITKFSYPGLLTALQYLTSVAGVWTLGKLGLLYHDPFNLQTAKKF 73
Query: 1031 LPAAVMFYISIFTNSELLLHADVDTFIVFRSAVPIFVAIGETLYLHQPWPSLKTWLSLST 852
PAA++FY++IFTN+ LL HA+VDTFIVFRS P+ VAI +T + QP PS T++SL T
Sbjct: 74 APAALVFYLAIFTNTHLLKHANVDTFIVFRSLTPLLVAIADTAFRKQPCPSKLTFVSLVT 133
Query: 851 ILGGSVIYVFTDNQFTVTAYTWAVAYLASMSIDFVYIKHVVMTIGLNTWGLVLYNNXXXX 672
ILGG+V YV TD+ F++TAY+WAVAYL +++ + VYIKH+V +GLNTWG VLYNN
Sbjct: 134 ILGGAVGYVMTDSGFSLTAYSWAVAYLVTITTEMVYIKHMVTKLGLNTWGFVLYNNLLSL 193
Query: 671 XXXXXXXLVMGE-FDQMKVDSSKVSNWLSFDVVLPVAXXXXXXXXXXXXXXXCRRAISAT 495
+ GE + S+ +W D + V+ R+AISAT
Sbjct: 194 IIAPVFWFLTGEHLSVFRAIESRGQSWFELDAFVAVSLSCVFGLLISFFGFAARKAISAT 253
Query: 494 GFTVLGIVNKLLTVVINLLIWDKHASFVGTIGLLICMSGGVLYQQSTT 351
FTV G+VNK LTV IN++IWDKHAS G + LL ++GGVLYQQS T
Sbjct: 254 AFTVTGVVNKFLTVAINVMIWDKHASSFGLVCLLFTLAGGVLYQQSVT 301
>sptr|Q84L08|Q84L08 GONST4 golgi nucleotide sugar transporter
(Fragment).
Length = 341
Score = 267 bits (683), Expect = 3e-70
Identities = 148/296 (50%), Positives = 184/296 (62%), Gaps = 1/296 (0%)
Frame = -1
Query: 1241 QQASIYGVAAGYCLSASLLSIINKWAIMKFPYPGALTALQYFTSVAGVLLCGQLKLIEHD 1062
+Q + + GY L +SLL++INK AI F YPG LTALQY T V L G+ LI HD
Sbjct: 10 KQLTTSSLVIGYALCSSLLAVINKLAITYFNYPGLLTALQYLTCTVAVYLLGKSGLINHD 69
Query: 1061 GLNLRTMWKFLPAAVMFYISIFTNSELLLHADVDTFIVFRSAVPIFVAIGETLYLHQPWP 882
T KFLPAA++FY++IFTN+ LL HA+VDTFIVFRS P+ VAI +T++ QP P
Sbjct: 70 PFTWDTAKKFLPAAIVFYLAIFTNTNLLRHANVDTFIVFRSLTPLLVAIADTVFRSQPLP 129
Query: 881 SLKTWLSLSTILGGSVIYVFTDNQFTVTAYTWAVAYLASMSIDFVYIKHVVMTIGLNTWG 702
S T+LSL IL G+V YV TD+ FT+TAY+WA+AYL +++ + VYIKH+V I LN WG
Sbjct: 130 SRLTFLSLVVILAGAVGYVATDSSFTLTAYSWALAYLVTITTEMVYIKHMVSNIKLNIWG 189
Query: 701 LVLYNNXXXXXXXXXXXLVMGEFDQM-KVDSSKVSNWLSFDVVLPVAXXXXXXXXXXXXX 525
LVLYNN + GEF ++ S N VA
Sbjct: 190 LVLYNNLLSLMIAPVFWFLTGEFTEVFAALSENRGNLFEPYAFSSVAASCVFGFLISYFG 249
Query: 524 XXCRRAISATGFTVLGIVNKLLTVVINLLIWDKHASFVGTIGLLICMSGGVLYQQS 357
R AISAT FTV G+VNK LTVVIN+LIWDKHA+ VG + LL + GGV YQQS
Sbjct: 250 FAARNAISATAFTVTGVVNKFLTVVINVLIWDKHATPVGLVCLLFTICGGVGYQQS 305
>sw|Q941R4|GON1_ARATH GDP-mannose transporter.
Length = 333
Score = 74.3 bits (181), Expect = 5e-12
Identities = 66/321 (20%), Positives = 136/321 (42%), Gaps = 9/321 (2%)
Frame = -1
Query: 1292 GTTERTGAWSNMLNTLMQQASIYGVAAGYCLSASLLSIINKWAIMKFPYPGALTALQY-- 1119
G T ++G + + +A + G+A YC+S+ + ++NK+ + + + + + Y
Sbjct: 14 GKTVKSGGDKPIPRKIHNRALLSGLA--YCISSCSMILVNKFVLSSYNFNAGIFLMLYQN 71
Query: 1118 FTSVAGVLLCGQLKLIEHDGLNLRTMWKFLPAAVMFYISIFTNSELLLHADVDTFIVFRS 939
F SV V+ + LI + L LR M + P V+F + T+ L + +V V ++
Sbjct: 72 FVSVIIVVGLSLMGLITTEPLTLRLMKVWFPVNVIFVGMLITSMFSLKYINVAMVTVLKN 131
Query: 938 AVPIFVAIGETLYLHQPWPSLKTWLSLSTILGGSVIYVFTDNQFTVTAYTWAVA------ 777
+ A+GE +YL + W +L ++ +V TD F Y W +A
Sbjct: 132 VTNVITAVGE-MYLFNKQHDNRVWAALFLMIISAVSGGITDLSFNAVGYAWQIANCFLTA 190
Query: 776 -YLASMSIDFVYIKHVVMTIGLNTWGLVLYNNXXXXXXXXXXXLVMGEFDQMKVDSSKVS 600
Y ++ K V + LN + +VL NN E D + + +
Sbjct: 191 SYSLTLRKTMDTAKQVTQSGNLNEFSMVLLNNTLSLPLGLLLSYFFNEMDYLY--QTPLL 248
Query: 599 NWLSFDVVLPVAXXXXXXXXXXXXXXXCRRAISATGFTVLGIVNKLLTVVINLLIWDKHA 420
SF +V+ ++ + AT ++++G +NK+ + +++++
Sbjct: 249 RLPSFWMVMTLSGLLGLAISFTSMWFLHQ--TGATTYSLVGSLNKIPLSIAGIVLFNVPT 306
Query: 419 SFVGTIGLLICMSGGVLYQQS 357
S + +L + GV++ ++
Sbjct: 307 SLQNSASILFGLVAGVVFARA 327
>sptr|Q9UTK8|Q9UTK8 Putative vanadate resistance protein.
Length = 345
Score = 65.1 bits (157), Expect = 3e-09
Identities = 62/309 (20%), Positives = 130/309 (42%), Gaps = 9/309 (2%)
Frame = -1
Query: 1265 SNMLNTLMQQASIYGVAAGYCLSASLLSIINKWAIMKFPYPG--ALTALQYFTSVAGVLL 1092
++MLN + + + V+ YC+++ L+++ NK+ + Y L +Q VA + +
Sbjct: 3 NHMLNRISKSPILPVVS--YCMASILMTLTNKYVLSSPGYNMNFLLLTVQSTVCVAAIGI 60
Query: 1091 CGQLKLIEHDGLNLRTMWKFLPAAVMFYISIFTNSELLLHADVDTFIVFRSAVPIFVAIG 912
+LK+I + + R + P + + I+T S+ L V + +F++ I +A G
Sbjct: 61 LKRLKVINYRDFDFREAKFWFPISFLLVAMIYTASKALQFLSVPVYTIFKNLTIIIIAYG 120
Query: 911 ETLYLHQPWPSLKTWLSLSTILGGSVIYVFTDNQFTVTA-------YTWAVAYLASMSID 753
E L+ +L T S ++ S++ + D Q + A Y W V + +
Sbjct: 121 EVLWFGGHVTAL-TLFSFGLMVLSSIVAAWADIQSSSFASQTLNSGYLWMVLNCLTNAAF 179
Query: 752 FVYIKHVVMTIGLNTWGLVLYNNXXXXXXXXXXXLVMGEFDQMKVDSSKVSNWLSFDVVL 573
+ ++ + + + YNN L ++ + + + F V++
Sbjct: 180 VLAMRKRIKLTNFRDFDTMFYNNLLSIPVLVICTLFTEDWSAENIAQNFPPD-AKFGVLM 238
Query: 572 PVAXXXXXXXXXXXXXXXCRRAISATGFTVLGIVNKLLTVVINLLIWDKHASFVGTIGLL 393
+A C R S+T ++++G +NKL + L+ +D +F +L
Sbjct: 239 AMAISGVSSVGISYTSAWCVRVTSSTTYSMVGALNKLPLAIAGLVFFDAPITFGSVTAIL 298
Query: 392 ICMSGGVLY 366
+ GV+Y
Sbjct: 299 LGFISGVVY 307
>sptr|Q9GNM8|Q9GNM8 LPG2 protein.
Length = 341
Score = 63.5 bits (153), Expect = 8e-09
Identities = 61/297 (20%), Positives = 130/297 (43%), Gaps = 4/297 (1%)
Frame = -1
Query: 1244 MQQASIYGVAAGYCLSASLLSIINKWAIMKFP--YPGALTALQYFTSVAGVLLCGQLKLI 1071
+ +A + + +C S S++ ++NK + + +P + LQ +V V L + I
Sbjct: 7 VMEAVLSVITYSFC-SVSMI-LVNKLIMNTYDMNFPFGILVLQTGGAVVIVALAKAARFI 64
Query: 1070 EHDGLNLRTMWKFLPAAVMFYISIFTNSELLLHADVDTFIVFRSAVPIFVAIGETLYLHQ 891
++ + ++LP ++F +FT+ + L V + ++ + +A+G+ +L+
Sbjct: 65 DYPAFSFDVAKQWLPLTLLFVAMLFTSMKSLGTMSVAAQTILKNLAVVLIALGDR-FLYG 123
Query: 890 PWPSLKTWLSLSTILGGSVIYVFTDNQFTVTAYTWAVAYLASMSIDFVYIKHVVMTIG-- 717
+ + S + ++ GS++ D T W + S +Y+K V+ ++
Sbjct: 124 KAQTPMVYFSFALMILGSLLGAKGDRWVTAWGLVWTFLNIVSTVSYTLYMKAVLGSVSNS 183
Query: 716 LNTWGLVLYNNXXXXXXXXXXXLVMGEFDQMKVDSSKVSNWLSFDVVLPVAXXXXXXXXX 537
+ +G V YNN +MG D M S+ + +F ++ +A
Sbjct: 184 IGRYGPVFYNNLLSLPFFL----IMGVGDMMPF-SAAIGETTTFGRLV-LAFSVLVSSVM 237
Query: 536 XXXXXXCRRAISATGFTVLGIVNKLLTVVINLLIWDKHASFVGTIGLLICMSGGVLY 366
C S T +V+G +NK+ + +L++ + + G +G+LI +S G LY
Sbjct: 238 TFSVFWCMSITSPTTMSVVGSLNKIPLTFLGMLVFHQFPTATGYLGILIALSAGFLY 294
>sptr|Q25266|Q25266 LPG2.
Length = 341
Score = 62.4 bits (150), Expect = 2e-08
Identities = 62/297 (20%), Positives = 130/297 (43%), Gaps = 4/297 (1%)
Frame = -1
Query: 1244 MQQASIYGVAAGYCLSASLLSIINKWAIMKFP--YPGALTALQYFTSVAGVLLCGQLKLI 1071
+ +A + + +C S S++ ++NK + + +P + LQ ++ V L + I
Sbjct: 7 VMEAVLAVITYSFC-SVSMI-LVNKLIMNTYDMNFPFGILVLQTGGALVIVALAKAARFI 64
Query: 1070 EHDGLNLRTMWKFLPAAVMFYISIFTNSELLLHADVDTFIVFRSAVPIFVAIGETLYLHQ 891
E+ + K+LP ++F +FT+ + L V + ++ + +A+G+ +L+
Sbjct: 65 EYPAFSFDVAKKWLPLTLLFVAMLFTSMKSLGTMSVAAQTILKNLAVVLIALGDK-FLYG 123
Query: 890 PWPSLKTWLSLSTILGGSVIYVFTDNQFTVTAYTWAVAYLASMSIDFVYIKHVVMTI--G 717
+ + S + ++ GS++ D T W + S +Y+K V+ ++
Sbjct: 124 KAQTPMVYFSFALMILGSLLGAKGDKWVTAWGLVWTFLNIVSTVSYTLYMKAVLGSVSNS 183
Query: 716 LNTWGLVLYNNXXXXXXXXXXXLVMGEFDQMKVDSSKVSNWLSFDVVLPVAXXXXXXXXX 537
+ +G V YNN L+MG D M S+ + + +F L +
Sbjct: 184 IGRYGPVFYNN----LLSLPFFLIMGVGDIMPF-SAAIGDTTTFG-KLVLTFSVLVSSVM 237
Query: 536 XXXXXXCRRAISATGFTVLGIVNKLLTVVINLLIWDKHASFVGTIGLLICMSGGVLY 366
C S T +V+G +NK+ + +L++ + + G +G++I +S G LY
Sbjct: 238 TFSVFWCMSITSPTTMSVVGSLNKIPLTFLGMLVFHQFPTATGYLGIMIALSAGFLY 294
>sptrnew|BAD03877|BAD03877 Hypothetical protein OJ1118_F01.31.
Length = 221
Score = 61.6 bits (148), Expect = 3e-08
Identities = 27/42 (64%), Positives = 34/42 (80%)
Frame = -1
Query: 476 IVNKLLTVVINLLIWDKHASFVGTIGLLICMSGGVLYQQSTT 351
+VNK LTV IN++IWDKHAS +G + LL ++GGVLYQQS T
Sbjct: 95 VVNKFLTVAINVMIWDKHASSIGLVCLLFTLAGGVLYQQSVT 136
Score = 48.9 bits (115), Expect = 2e-04
Identities = 25/43 (58%), Positives = 28/43 (65%)
Frame = -1
Query: 1157 KFPYPGALTALQYFTSVAGVLLCGQLKLIEHDGLNLRTMWKFL 1029
KF YPG LTALQY TSVAGV +L L+ HD N +T K L
Sbjct: 3 KFSYPGLLTALQYLTSVAGVWTLAKLGLLYHDPFNFQTAKKSL 45
>sptrnew|BAD03089|BAD03089 Hypothetical protein OJ1484_G09.115.
Length = 221
Score = 61.6 bits (148), Expect = 3e-08
Identities = 27/42 (64%), Positives = 34/42 (80%)
Frame = -1
Query: 476 IVNKLLTVVINLLIWDKHASFVGTIGLLICMSGGVLYQQSTT 351
+VNK LTV IN++IWDKHAS +G + LL ++GGVLYQQS T
Sbjct: 95 VVNKFLTVAINVMIWDKHASSIGLVCLLFTLAGGVLYQQSVT 136
Score = 48.9 bits (115), Expect = 2e-04
Identities = 25/43 (58%), Positives = 28/43 (65%)
Frame = -1
Query: 1157 KFPYPGALTALQYFTSVAGVLLCGQLKLIEHDGLNLRTMWKFL 1029
KF YPG LTALQY TSVAGV +L L+ HD N +T K L
Sbjct: 3 KFSYPGLLTALQYLTSVAGVWTLAKLGLLYHDPFNFQTAKKSL 45
>sptrnew|BAD09390|BAD09390 Putative GDP-Mannose transporter.
Length = 350
Score = 58.9 bits (141), Expect = 2e-07
Identities = 46/201 (22%), Positives = 91/201 (45%), Gaps = 10/201 (4%)
Frame = -1
Query: 1256 LNTLMQQASIYGVAAGYCLSASLLSIINKWAIMKFPYPGALTALQYFTSVAGVLLCGQLK 1077
L + QA + G+A YC+S+ + ++NK+ + + + + + Y ++ V + L
Sbjct: 91 LPKIQNQALLSGLA--YCISSCSMILVNKYILSGYGFSAGIFLMLY-QNIVSVTIVSTLS 147
Query: 1076 L---IEHDGLNLRTMWKFLPAAVMFYISIFTNSELLLHADVDTFIVFRSAVPIFVAIGET 906
L I + L + + +LP ++F + T+ L + +V + ++ + A GET
Sbjct: 148 LSGVIPTEPLTWKLIKVWLPVNIIFVGMLITSMFSLKYINVAMLTILKNVANVLTASGET 207
Query: 905 LYLHQPWPSLKTWLSLSTILGGSVIYVFTDNQFTVTAYTWAVA---YLASMSIDFVYI-- 741
Y + + W+SL+ ++ ++ TD F YTW + AS S+ ++
Sbjct: 208 -YFFKKQHDRQVWISLTLMIISAIAGGITDLSFNAIGYTWQILNCFLTASYSLTLRHVMD 266
Query: 740 --KHVVMTIGLNTWGLVLYNN 684
K + LN +VL NN
Sbjct: 267 SAKQATKSGNLNELSMVLLNN 287
>sptr|Q96WN8|Q96WN8 Golgi GDP-mannose transporter.
Length = 371
Score = 58.5 bits (140), Expect = 3e-07
Identities = 67/334 (20%), Positives = 133/334 (39%), Gaps = 15/334 (4%)
Frame = -1
Query: 1322 SKEDTSPEGSGTTERTGAWSNMLNTLMQQASIYGVAAGYCLSASLLSIINKWAIMKFPY- 1146
SK TS SG+ L T + + +AA YCLS+ L+++ NK+ + F +
Sbjct: 36 SKHSTSSSSSGS----------LATRISNSGPISIAA-YCLSSILMTVTNKYVLSGFSFN 84
Query: 1145 -PGALTALQYFTSVAGVLLCGQLKLIEHDGLNLRTMWKFLPAAVMFYISIFTNSELLLHA 969
L A+Q + + L +I + N K+ P A + I+T+S+ L +
Sbjct: 85 LNFFLLAVQSIVCIVTIGSLKSLNIITYRQFNKDEAKKWSPIAFLLVAMIYTSSKALQYL 144
Query: 968 DVDTFIVFRSAVPIFVAIGETLYLHQPWPSLKTWLSLSTIL---GGSVIYVFTDNQFTVT 798
+ + +F++ I +A GE ++ + T ++LS+ L SVI + DN +
Sbjct: 145 SIPVYTIFKNLTIILIAYGEVIW----FGGKVTTMALSSFLLMVLSSVIAYYGDNAAVKS 200
Query: 797 ---------AYTWAVA-YLASMSIDFVYIKHVVMTIGLNTWGLVLYNNXXXXXXXXXXXL 648
Y W + AS + + K + +T + + YNN
Sbjct: 201 HDDAFALYLGYFWMLTNCFASAAFVLIMRKRIKLT-NFKDFDTMYYNNLLSIPILLICSF 259
Query: 647 VMGEFDQMKVDSSKVSNWLSFDVVLPVAXXXXXXXXXXXXXXXCRRAISATGFTVLGIVN 468
+ ++ V + ++ + + C R S+T ++++G +N
Sbjct: 260 IFEDWSSANVSLNFPAD-NRVTTITAMILSGASSVGISYCSAWCVRVTSSTTYSMVGALN 318
Query: 467 KLLTVVINLLIWDKHASFVGTIGLLICMSGGVLY 366
KL + L+ ++ +F + + G++Y
Sbjct: 319 KLPIALSGLIFFEAAVNFWSVSSIFVGFGAGLVY 352
>sptr|Q9BIR6|Q9BIR6 Lipophosphoglycan biosynthetic protein.
Length = 341
Score = 58.2 bits (139), Expect = 3e-07
Identities = 58/297 (19%), Positives = 126/297 (42%), Gaps = 4/297 (1%)
Frame = -1
Query: 1244 MQQASIYGVAAGYCLSASLLSIINKWAIMKFP--YPGALTALQYFTSVAGVLLCGQLKLI 1071
+ +A + + +C S S++ ++NK + + +P + LQ ++ V L + +
Sbjct: 7 VMEAILAVITYSFC-SVSMI-LVNKLIMNTYDMNFPFGILVLQTGGALVIVALAKAARFV 64
Query: 1070 EHDGLNLRTMWKFLPAAVMFYISIFTNSELLLHADVDTFIVFRSAVPIFVAIGETLYLHQ 891
E+ + K+LP ++F +FT+ + L V + ++ I +A+G+ +L+
Sbjct: 65 EYPAFSFDVAKKWLPLTLLFVAMLFTSMKSLATMSVAAQTILKNLAVILIALGDK-FLYG 123
Query: 890 PWPSLKTWLSLSTILGGSVIYVFTDNQFTVTAYTWAVAYLASMSIDFVYIKHVVMTI--G 717
+ + S + ++ GS + D T W + + S +Y+K V+ ++
Sbjct: 124 KAQTPMVYFSFALMILGSFLGAKGDKWVTAWGLIWTLLNIVSTVSYTLYMKAVLGSVSNS 183
Query: 716 LNTWGLVLYNNXXXXXXXXXXXLVMGEFDQMKVDSSKVSNWLSFDVVLPVAXXXXXXXXX 537
+ +G V YNN L+MG + M ++ +VL +
Sbjct: 184 IGRYGPVFYNN----LLSLPFFLIMGVGEIMPFSAAIGETTTLGKLVLTFSVLVSSVMTF 239
Query: 536 XXXXXXCRRAISATGFTVLGIVNKLLTVVINLLIWDKHASFVGTIGLLICMSGGVLY 366
S T +V+G +NK+ + +L + + + G +G+++ +S G LY
Sbjct: 240 SVFWCMS--ITSPTTMSVVGSLNKIPLTFLGMLAFHQFPTATGYLGIMVALSAGFLY 294
>sptr|Q7S227|Q7S227 Hypothetical protein.
Length = 338
Score = 55.8 bits (133), Expect = 2e-06
Identities = 63/292 (21%), Positives = 120/292 (41%), Gaps = 9/292 (3%)
Frame = -1
Query: 1214 AGYCLSASLLSIINKWAIMKFPYPGALTALQYFTSVAG---VLLCGQLKLIEHDGLNLRT 1044
A Y L L+I NK + KF YP LTAL ++ G +LL G+ L + L+L+
Sbjct: 45 AVYFLCNISLTIYNKLILGKFSYPWLLTALHAGSASIGCYILLLQGRFTLTK---LSLQQ 101
Query: 1043 MWKFLPAAVMFYISIFTNSELLLHADVDTFIVFRSAVPIFVAIGETLYLHQPWPSLKTWL 864
+++F ++I T++ L + + RS P F + + +P T+L
Sbjct: 102 NIVLFLFSILFTVNIATSNVSLAMVSIPFHQIMRSTCPFFAVLIYRFRYGRSYPR-DTYL 160
Query: 863 SLSTILGGSVIYVFTDNQFTVTAYTWAVAYLASMSIDFVYIKHV----VMT--IGLNTWG 702
SL ++ G + + D FT + L + + +K V +MT + L+
Sbjct: 161 SLIPLILGVGLATYGDYYFTAAGF-----LLTFLGVILAVVKTVATNRIMTGALALSPLE 215
Query: 701 LVLYNNXXXXXXXXXXXLVMGEFDQMKVDSSKVSNWLSFDVVLPVAXXXXXXXXXXXXXX 522
+L + + GE K + + S ++L +A
Sbjct: 216 TLLRMSPLACAQALVCAIASGELAGFKEQNPEGP---SGALILTLAGNGLLAFCLNYSSF 272
Query: 521 XCRRAISATGFTVLGIVNKLLTVVINLLIWDKHASFVGTIGLLICMSGGVLY 366
+ A TV G + + LT+++ ++++ F+ +G++I ++G Y
Sbjct: 273 STNKVAGAVTMTVCGNIKQCLTILLGIVLFGVKVGFLNGLGMVIALAGAAWY 324
>sptr|Q8AV24|Q8AV24 UGNT.
Length = 297
Score = 55.5 bits (132), Expect = 2e-06
Identities = 54/254 (21%), Positives = 99/254 (38%), Gaps = 1/254 (0%)
Frame = -1
Query: 1124 QYFTSVAGVLLCGQLKLIEHDGLNLRTMWKFLPAAVMFYISIFTNSELLLHADVDTFIVF 945
Q FT+V + +K ++ + + K P +++ + T + F V
Sbjct: 16 QMFTTVVVLYAAKMIKTVQFQDFDRSVLIKIFPLPLLYVGNHITGLASTKKLSLPMFTVL 75
Query: 944 RSAVPIFVAIGETLYLHQPWPSLKTWLSLSTILGGSVIYVFTDNQFTVTAYTWAVAYLAS 765
R + I E L + +P + S+ I+ G+++ +D F V YT+ + A
Sbjct: 76 RKFTILMTMILEVYILRKRFPKRLVY-SVMAIVFGAMVAASSDLAFDVQGYTFILLNEAF 134
Query: 764 MSIDFVYIKHVVMTIGLNTWGLVLYNNXXXXXXXXXXXLVMGEFDQMKVDSSKVSNWLSF 585
+ VY K + T GL +G++ YN ++ F + + ++W+
Sbjct: 135 TAASNVYTKKNLGTEGLGKYGVLFYN---ALIHSFVPTILASAFTGDLHKAVEFADWVKA 191
Query: 584 DVVLPVAXXXXXXXXXXXXXXXCRRAISATGFTVLGIVNKLLTVVINLLI-WDKHASFVG 408
V C SA T++G + + I + + D S++
Sbjct: 192 PFVFSFLMSCFMGFVLMYSIVLCSYYNSALTTTIVGAIKNVAVAYIGIFVGGDYLFSWLN 251
Query: 407 TIGLLICMSGGVLY 366
IGL ICMSGG+ Y
Sbjct: 252 FIGLSICMSGGLAY 265
>sptr|Q7SBC5|Q7SBC5 Hypothetical protein.
Length = 392
Score = 55.5 bits (132), Expect = 2e-06
Identities = 59/313 (18%), Positives = 124/313 (39%), Gaps = 27/313 (8%)
Frame = -1
Query: 1208 YCLSASLLSIINKWAI------MKFPYPGALTALQYFTSVAGVLLCGQLKLIEH-DGLNL 1050
YCLS+ ++++NK+ + + F Y A+Q A +L+C QL + ++ +
Sbjct: 61 YCLSSISMTVVNKYVVSGSEWNLNFFY----LAVQSLVCTAAILICKQLGMFQNLAAFDS 116
Query: 1049 RTMWKFLPAAVMFYISIFTNSELLLHADVDTFIVFRSAVPIFVAIGETLYLHQPWPSLKT 870
K+ P +++ I+T+++ L V + +F++ I VA GE L+ +
Sbjct: 117 TKAKKWFPISLLLVGMIYTSTKALQFLSVPVYTIFKNLTIIVVAYGEVLWFGGSVTPM-A 175
Query: 869 WLSLSTILGGSVIYVFTDNQFTV-----------------TAYTWAVAYLASMSIDFVYI 741
LS ++ SVI + D Q V Y W + + + +
Sbjct: 176 LLSFGLMVLSSVIAAWADIQAAVEGVGHTAEATDAISTLNAGYAWMGMNVFCTAAYLLGM 235
Query: 740 KHVVMTIGLNTWGLVLYNNXXXXXXXXXXXLVMGEFDQMKVDSSKVSNW---LSFDVVLP 570
+ V+ + + + YNN L+ ++ + + + N+ + +
Sbjct: 236 RKVIKKMNFKDYDTMFYNNLLTIPVLIVFSLLFEDWS----NDNLIKNFPVETRNSLFIG 291
Query: 569 VAXXXXXXXXXXXXXXXCRRAISATGFTVLGIVNKLLTVVINLLIWDKHASFVGTIGLLI 390
+ C R S+T ++++G +NKL + L+ +D +F + +
Sbjct: 292 MIYSGLAAIFISYCSAWCIRVTSSTTYSMVGALNKLPLAISGLIFFDAPVTFGSVTAIFV 351
Query: 389 CMSGGVLYQQSTT 351
G++Y S T
Sbjct: 352 GFVSGLVYTWSKT 364
>sptrnew|AAM76201|AAM76201 RH08077p.
Length = 373
Score = 55.1 bits (131), Expect = 3e-06
Identities = 62/293 (21%), Positives = 115/293 (39%), Gaps = 3/293 (1%)
Frame = -1
Query: 1223 GVAAGYCLSASLLSIINKWAIMKFPYPGAL-TALQYFTSVAGVLLCGQ-LKLIEHDGLNL 1050
G A Y LS+ +++++NK + + +P L +L T+ VL G+ LKL+ L
Sbjct: 65 GSALFYGLSSFMITVVNKTVLTSYHFPSFLFLSLGQLTASIVVLGMGKRLKLVNFPPLQR 124
Query: 1049 RTMWKFLPAAVMFYISIFTNSELLLHADVDTFIVFRSAVPIFVAIGETLYLHQPWPSLKT 870
T K P ++F ++ + F R + + E L PS
Sbjct: 125 NTFAKIFPLPLIFLGNMMFGLGGTKTLSLPMFAALRRFSILMTMLLELKILGLR-PSNAV 183
Query: 869 WLSLSTILGGSVIYVFTDNQFTVTAYTWAVAYLASMSIDFVYIKHVVMTIGLNTWGLVLY 690
+S+ ++GG+++ D F + Y + + A + + VY+K + T + +GL+ Y
Sbjct: 184 QVSVYAMIGGALLAASDDLSFNMRGYIYVMITNALTASNGVYVKKKLDTSEIGKYGLMYY 243
Query: 689 NNXXXXXXXXXXXLVMGEFDQMKVDSSKVSNWLSFDVVLPVAXXXXXXXXXXXXXXXCRR 510
N+ V G DQ + W V+ C +
Sbjct: 244 NSLFMFLPALALNYVTGNLDQ----ALNFEQWNDSVFVVQFLLSCVMGFILSYSTILCTQ 299
Query: 509 AISATGFTVLGIVNKLLTVVINLLIWDKHA-SFVGTIGLLICMSGGVLYQQST 354
SA T++G + + + + I + S++ IG+ I + +LY T
Sbjct: 300 FNSALTTTIVGCLKNICVTYLGMFIGGDYVFSWLNCIGINISVLASLLYTYVT 352
>sw|Q95YI5|US74_DROME UDP-sugar transporter UST74c (Fringe connection
protein).
Length = 373
Score = 55.1 bits (131), Expect = 3e-06
Identities = 62/293 (21%), Positives = 115/293 (39%), Gaps = 3/293 (1%)
Frame = -1
Query: 1223 GVAAGYCLSASLLSIINKWAIMKFPYPGAL-TALQYFTSVAGVLLCGQ-LKLIEHDGLNL 1050
G A Y LS+ +++++NK + + +P L +L T+ VL G+ LKL+ L
Sbjct: 65 GSALFYGLSSFMITVVNKTVLTSYHFPSFLFLSLGQLTASIVVLGMGKRLKLVNFPPLQR 124
Query: 1049 RTMWKFLPAAVMFYISIFTNSELLLHADVDTFIVFRSAVPIFVAIGETLYLHQPWPSLKT 870
T K P ++F ++ + F R + + E L PS
Sbjct: 125 NTFAKIFPLPLIFLGNMMFGLGGTKTLSLPMFAALRRFSILMTMLLELKILGLR-PSNAV 183
Query: 869 WLSLSTILGGSVIYVFTDNQFTVTAYTWAVAYLASMSIDFVYIKHVVMTIGLNTWGLVLY 690
+S+ ++GG+++ D F + Y + + A + + VY+K + T + +GL+ Y
Sbjct: 184 QVSVYAMIGGALLAASDDLSFNMRGYIYVMITNALTASNGVYVKKKLDTSEIGKYGLMYY 243
Query: 689 NNXXXXXXXXXXXLVMGEFDQMKVDSSKVSNWLSFDVVLPVAXXXXXXXXXXXXXXXCRR 510
N+ V G DQ + W V+ C +
Sbjct: 244 NSLFMFLPALALNYVTGNLDQ----ALNFEQWNDSVFVVQFLLSCVMGFILSYSTILCTQ 299
Query: 509 AISATGFTVLGIVNKLLTVVINLLIWDKHA-SFVGTIGLLICMSGGVLYQQST 354
SA T++G + + + + I + S++ IG+ I + +LY T
Sbjct: 300 FNSALTTTIVGCLKNICVTYLGMFIGGDYVFSWLNCIGINISVLASLLYTYVT 352
>sptr|Q84L09|Q84L09 GONST2 golgi nucleotide sugar transporter
(Fragment).
Length = 375
Score = 55.1 bits (131), Expect = 3e-06
Identities = 60/297 (20%), Positives = 124/297 (41%), Gaps = 11/297 (3%)
Frame = -1
Query: 1214 AGYCLSASLLSIINKWAIMKFPYPGALTALQYFTSVA----GVLLCGQLKLIEHDGLNLR 1047
A YC+S+ + I+NK + + + ++ + Y ++ VL + +E L
Sbjct: 83 AAYCISSCSMIILNKIVLSSYNFNAGVSLMLYQNLISCLVVAVLDISGVVSVEKFNWKLI 142
Query: 1046 TMWKFLPAAVMFYISIFTNSELLLHADVDTFIVFRSAVPIFVAIGETLYLHQPWPSLKTW 867
+W +P V+F + + L + +V + ++A I IGE +Y+ + + K W
Sbjct: 143 RVW--MPVNVIFVGMLVSGMYSLKYINVAMVTILKNATNILTGIGE-VYMFRKRQNNKVW 199
Query: 866 LSLSTILGGSVIYVFTDNQFTVTAYTWAVA---YLASMSIDFVYI----KHVVMTIGLNT 708
++ ++ ++ TD F YTW +A AS S+ + K + LN
Sbjct: 200 AAMFMMIISAISGGITDLTFDAVGYTWQLANCFLTASYSLTLRRVMDKAKQSTKSGSLNE 259
Query: 707 WGLVLYNNXXXXXXXXXXXLVMGEFDQMKVDSSKVSNWLSFDVVLPVAXXXXXXXXXXXX 528
+VL NN +++GE+ V S+ V+ F VV +
Sbjct: 260 VSMVLLNNLLSIPFGIILIILLGEW--RYVISTDVTKDSMFWVVATASGFLGLAISFTSM 317
Query: 527 XXXCRRAISATGFTVLGIVNKLLTVVINLLIWDKHASFVGTIGLLICMSGGVLYQQS 357
+ T ++++G +NK+ + L++++ S +L + GV++ ++
Sbjct: 318 WFLHQ--TGPTTYSLVGSLNKVPISLAGLVLFNVPLSLPNLFSILFGLFAGVVFARA 372
>sptr|Q94B65|Q94B65 Hypothetical protein.
Length = 323
Score = 54.7 bits (130), Expect = 4e-06
Identities = 55/285 (19%), Positives = 116/285 (40%), Gaps = 1/285 (0%)
Frame = -1
Query: 1217 AAGYCLSASLLSIINKWAIMKFPYPGALTALQYFTSVAGVLLCGQLKLIEHDGLNLRTMW 1038
A Y +++ + INK IM++P+ + LQ + + ++ G+++ T
Sbjct: 18 AVSYGIASMAMVFINKAVIMQYPHSMTVLTLQQLATSLLIHFGRRMGYTRAKGIDMATAK 77
Query: 1037 KFLPAAVMFYISIFTNSELLLHADVDTFIVFRSAVPIFVAIGETLYLHQPWPSLKTWLSL 858
K LP ++ + ++ L ++ +I + P+ V I L+ + P+ + LS+
Sbjct: 78 KLLPVSIFYNANVAFALASLKGVNIPMYIAIKRLTPLAVLISGVLF-GKGKPTTQVALSV 136
Query: 857 STILGGSVIYVFTDNQFTVTAYTWAVAYLASMSIDFVYIKHVVMTIGLNTWGLVLYNNXX 678
G VI D F + Y A+ + ++ V ++ GL++ ++ YN+
Sbjct: 137 LLTAAGCVIAALGDFSFDLFGYGLALTSVFFQTMYLVLVEKSGAEDGLSSIEIMFYNSFL 196
Query: 677 XXXXXXXXXLVMGEFDQMKVDSSKVSNWLSFDVVLPVAXXXXXXXXXXXXXXXCRRAISA 498
+V GEF ++L F V+L ++ SA
Sbjct: 197 SLPFLSILIIVTGEFPNSLSLLLAKCSYLPFLVILILSLVMGIVLNFTMFLCTI--VNSA 254
Query: 497 TGFTVLGIVNKLLTVVIN-LLIWDKHASFVGTIGLLICMSGGVLY 366
T++G++ + + + +L+ + GL++ +GGV Y
Sbjct: 255 LTTTIVGVLKGVGSTTLGFVLLGGVEVHALNVSGLVVNTAGGVWY 299
>sw|P40107|GOG5_YEAST GDP-mannose transporter.
Length = 337
Score = 53.5 bits (127), Expect = 8e-06
Identities = 51/301 (16%), Positives = 118/301 (39%), Gaps = 20/301 (6%)
Frame = -1
Query: 1208 YCLSASLLSIINKWAI--MKFPYPGALTALQYFTSVAGVLLCGQLKLIEHDGLNLRTMWK 1035
YC S+ L+++ NK+ + F + +Q +++ L + LN
Sbjct: 28 YCGSSILMTVTNKFVVNLKDFNMNFVMLFVQSLVCTITLIILRILGYAKFRSLNKTDAKN 87
Query: 1034 FLPAAVMFYISIFTNSELLLHADVDTFIVFRSAVPIFVAIGETLYLHQPWPSLKTWLSLS 855
+ P + + + I+T+S+ L + V + +F++ I +A GE L+ S++ L
Sbjct: 88 WFPISFLLVLMIYTSSKALQYLAVPIYTIFKNLTIILIAYGEVLFFGGSVTSMELSSFLL 147
Query: 854 TILGGSVIYVFTDNQFTVT------------------AYTWAVAYLASMSIDFVYIKHVV 729
+L SV+ + D Q Y W + ++ + ++ +
Sbjct: 148 MVLS-SVVATWGDQQAVAAKAASLAEGAAGAVASFNPGYFWMFTNCITSALFVLIMRKRI 206
Query: 728 MTIGLNTWGLVLYNNXXXXXXXXXXXLVMGEFDQMKVDSSKVSNWLSFDVVLPVAXXXXX 549
+ + YNN + ++ + + ++ ++ L+ ++ VA
Sbjct: 207 KLTNFKDFDTMFYNNVLALPILLLFSFCVEDWSSVNLTNNFSNDSLTAMIISGVASVGIS 266
Query: 548 XXXXXXXXXXCRRAISATGFTVLGIVNKLLTVVINLLIWDKHASFVGTIGLLICMSGGVL 369
R S+T ++++G +NKL + L+ +D +F+ + + I G++
Sbjct: 267 YCSGWCV-----RVTSSTTYSMVGALNKLPIALSGLIFFDAPRNFLSILSIFIGFLSGII 321
Query: 368 Y 366
Y
Sbjct: 322 Y 322
>sptr|Q96X08|Q96X08 GDP-mannose transporter.
Length = 324
Score = 53.1 bits (126), Expect = 1e-05
Identities = 55/296 (18%), Positives = 121/296 (40%), Gaps = 15/296 (5%)
Frame = -1
Query: 1208 YCLSASLLSIINKWAIMKFPYPGALTALQYFTSVAGVLLCGQLKLI---EHDGLNLRTMW 1038
YC S+ L+++ NK+ + + + + S+ + LK+ ++ LNL +
Sbjct: 25 YCASSILMTVTNKFVVNTDGF-NMFFVMLFAQSLVCTMCLMVLKMFGYAKYRPLNLIDVK 83
Query: 1037 KFLPAAVMFYISIFTNSELLLHADVDTFIVFRSAVPIFVAIGETLYLHQPWPSLK----T 870
+LP + + IFT+++ L + V + +F++ I +A GE L+ ++
Sbjct: 84 NWLPISFLLVFMIFTSAKALKYMPVPIYTIFKNLTIILIAYGEVLFFGGSVTPMELSSFI 143
Query: 869 WLSLSTIL-------GGSVIYVFTDNQFTVTAYTWA-VAYLASMSIDFVYIKHVVMTIGL 714
+ LS+++ + +N Y W + + S S + K + +T
Sbjct: 144 LMVLSSVVASLGDQQAAKIAQPLANNSILSPEYYWMFLNCICSASFVLIMRKRIKLT-NF 202
Query: 713 NTWGLVLYNNXXXXXXXXXXXLVMGEFDQMKVDSSKVSNWLSFDVVLPVAXXXXXXXXXX 534
+ + YNN + ++ S ++ S + + +
Sbjct: 203 KDYDTMFYNNALALPILLGFSFLSEDW-----SSENLAQNFSGESLSAMIISGMTSVGIS 257
Query: 533 XXXXXCRRAISATGFTVLGIVNKLLTVVINLLIWDKHASFVGTIGLLICMSGGVLY 366
C RA S+T ++++G +NKL + L+ +D +F+ + + I + G+LY
Sbjct: 258 YCSGWCVRATSSTTYSMVGALNKLPIALAGLIFFDAPRNFLSIMSIFIGFASGLLY 313
>sptr|Q7Q9J8|Q7Q9J8 AgCP15152 (Fragment).
Length = 576
Score = 51.6 bits (122), Expect = 3e-05
Identities = 53/291 (18%), Positives = 118/291 (40%), Gaps = 3/291 (1%)
Frame = -1
Query: 1217 AAGYCLSASLLSIINKWAIMKFPYPG--ALTALQYFTSVAGVLLCGQLKLIEHDGLNLRT 1044
A Y LS+ L++++NK + + +P L+ Q S+ + + +L+L+++ +
Sbjct: 66 AVFYGLSSFLITVVNKTVLTSYRFPSFLVLSLGQLTASIVVLFVAKRLQLVKYPDFSRDI 125
Query: 1043 MWKFLPAAVMFYISIFTNSELLLHADVDTFIVFRSAVPIFVAIGETLYLHQPWPSLKTWL 864
+ P +++ ++ + F R + + E L L P+L +
Sbjct: 126 ARRIFPLPLIYLGNMMFGLGGTQALSLPMFAALRRFSILLTMLLELLVLGIR-PTLAVKV 184
Query: 863 SLSTILGGSVIYVFTDNQFTVTAYTWAVAYLASMSIDFVYIKHVVMTIGLNTWGLVLYNN 684
S+ ++GG+++ D F + Y + + + + VY+K + T + +GL+ YN+
Sbjct: 185 SVFAMVGGALMAALDDLSFNLQGYMYVMITNTLTAANGVYMKKKLDTADMGKYGLMYYNS 244
Query: 683 XXXXXXXXXXXLVMGEFDQMKVDSSKVSNWLSFDVVLPVAXXXXXXXXXXXXXXXCRRAI 504
+ G+ D+ + + W V+ C +
Sbjct: 245 LFMILPALVGTWLAGDIDR----AWQYEGWNDPFFVVQFLLSCVMGFILSYSVILCTQHN 300
Query: 503 SATGFTVLGIVNKLLTVVINLLIWDKHA-SFVGTIGLLICMSGGVLYQQST 354
SA T++G + + I + I + S + +G+ I ++G +LY T
Sbjct: 301 SALTTTIVGCLKNISVTYIGMFIGGDYVFSLLNALGINISVAGSLLYTYVT 351
>sptr|Q9SB76|Q9SB76 Hypothetical protein.
Length = 296
Score = 50.8 bits (120), Expect = 5e-05
Identities = 53/272 (19%), Positives = 110/272 (40%), Gaps = 1/272 (0%)
Frame = -1
Query: 1178 INKWAIMKFPYPGALTALQYFTSVAGVLLCGQLKLIEHDGLNLRTMWKFLPAAVMFYISI 999
INK IM++P+ + LQ + + ++ G+++ T K LP ++ + ++
Sbjct: 4 INKAVIMQYPHSMTVLTLQQLATSLLIHFGRRMGYTRAKGIDMATAKKLLPVSIFYNANV 63
Query: 998 FTNSELLLHADVDTFIVFRSAVPIFVAIGETLYLHQPWPSLKTWLSLSTILGGSVIYVFT 819
L ++ +I + P+ V I L+ + P+ + LS+ G VI
Sbjct: 64 AFALASLKGVNIPMYIAIKRLTPLAVLISGVLF-GKGKPTTQVALSVLLTAAGCVIAALG 122
Query: 818 DNQFTVTAYTWAVAYLASMSIDFVYIKHVVMTIGLNTWGLVLYNNXXXXXXXXXXXLVMG 639
D F + Y A+ + ++ V ++ GL++ ++ YN+ +V G
Sbjct: 123 DFSFDLFGYGLALTSVFFQTMYLVLVEKSGAEDGLSSIEIMFYNSFLSLPFLSILIIVTG 182
Query: 638 EFDQMKVDSSKVSNWLSFDVVLPVAXXXXXXXXXXXXXXXCRRAISATGFTVLGIVNKLL 459
EF ++L F V+L ++ SA T++G++ +
Sbjct: 183 EFPNSLSLLLAKCSYLPFLVILILSLVMGIVLNFTMFLCTI--VNSALTTTIVGVLKGVG 240
Query: 458 TVVIN-LLIWDKHASFVGTIGLLICMSGGVLY 366
+ + +L+ + GL++ +GGV Y
Sbjct: 241 STTLGFVLLGGVEVHALNVSGLVVNTAGGVWY 272
>sptr|Q7SXZ5|Q7SXZ5 Hypothetical protein.
Length = 363
Score = 47.8 bits (112), Expect = 5e-04
Identities = 57/292 (19%), Positives = 115/292 (39%), Gaps = 4/292 (1%)
Frame = -1
Query: 1217 AAGYCLSASLLSIINKWAIMKFPYPG--ALTALQYFTSVAGVLLCGQLKLIEHDGLNLRT 1044
AA Y +S+ L+ ++NK + + +P AL Q F +V + L +I +
Sbjct: 38 AAFYGISSFLIVVVNKSVLTNYRFPSSLALGIGQMFATVVVLRGAKALNMISFPDFDWHV 97
Query: 1043 MWKFLPAAVMFYISIFTNSELLLHADVDTFIVFRSAVPIFVAIGETLYLHQPWP-SLKTW 867
+K P +++ + T ++ F V R +F + E L Q + S+K
Sbjct: 98 AYKVFPLPLLYVGNQLTGLFGTKQLNLPMFTVLRRFSILFTMLFEGYLLKQKFSWSIKA- 156
Query: 866 LSLSTILGGSVIYVFTDNQFTVTAYTWAVAYLASMSIDFVYIKHVVMTIGLNTWGLVLYN 687
++ T++ G+ + +D F + Y + + + Y+K + + L +GL+ YN
Sbjct: 157 -TVFTMILGAFVAASSDLAFDLQGYVFITLNNILTAANGAYMKQKLDSKELGKYGLLYYN 215
Query: 686 NXXXXXXXXXXXLVMGEFDQMKVDSSKVSNWLSFDVVLPVAXXXXXXXXXXXXXXXCRRA 507
G+ + + + W + V+ C
Sbjct: 216 ALLMIIPTVVLAYFTGDVQK----TLECEVWADYFFVIQFVLSCVMGFILMYSIMLCTHY 271
Query: 506 ISATGFTVLGIVNKLLTVVINLLI-WDKHASFVGTIGLLICMSGGVLYQQST 354
SA T++G + +L I ++ D S++ +GL I ++G ++Y T
Sbjct: 272 NSALTTTIVGCIKNILVTYIGMVFGGDYIFSWMNFVGLNISIAGSLVYSYIT 323
>sptr|Q94GE9|Q94GE9 Putative lipophosphoglycan biosynthetic.
Length = 368
Score = 47.4 bits (111), Expect = 6e-04
Identities = 44/211 (20%), Positives = 91/211 (43%), Gaps = 17/211 (8%)
Frame = -1
Query: 1265 SNMLNTLMQ-QASIYGVAAG-------YCLSASLLSIINKWAIMKFPYPGALTALQY--F 1116
S++ ++L+Q Q S +G G YC+++ + ++NK + + + ++ + Y
Sbjct: 58 SDVFDSLIQKQQSKWGNKTGPLLSGICYCIASCSMILLNKVVLSNYNFNAGISLMLYQNL 117
Query: 1115 TSVAGVLLCGQLKLIEHDGLNLRTMWKFLPAAVMFYISIFTNSELLLHADVDTFIVFRSA 936
SV +L+ +I + L + + ++P ++F + T L + +V + ++
Sbjct: 118 ISVIILLVLELFGVISTEKLTWKLIKVWIPVNLIFVGMLVTGMYSLKYINVAMVTILKNM 177
Query: 935 VPIFVAIGETLYLHQPWPSLKTWLSLSTILGGSVIYVFTDNQFTVTAYTW-------AVA 777
I A+GE +Y+ + + K W +L ++ +V TD F Y W
Sbjct: 178 TNILTAVGE-IYIFRKGQNKKVWAALCLMVISAVCGGITDLSFHPVGYMWQLFNCFLTAG 236
Query: 776 YLASMSIDFVYIKHVVMTIGLNTWGLVLYNN 684
Y ++ K + LN +VL NN
Sbjct: 237 YSLTLRRVMDVAKQSTKSGSLNEVSMVLLNN 267
>sptr|O64909|O64909 Glucose-6-phosphate/phosphate-translocator
precursor.
Length = 387
Score = 47.0 bits (110), Expect = 8e-04
Identities = 54/279 (19%), Positives = 102/279 (36%), Gaps = 1/279 (0%)
Frame = -1
Query: 1190 LLSIINKWAIMKFPYPGALTALQYFTSVAGVLLCGQLKLIEHDGLNLRTMWKFL-PAAVM 1014
+ +I NK + FPYP + L A +L +L+E +L WK L P AV
Sbjct: 109 IFNIYNKKVLNAFPYPWLTSTLSLACGSAMMLFSWATRLVEAPKTDL-DFWKVLFPVAVA 167
Query: 1013 FYISIFTNSELLLHADVDTFIVFRSAVPIFVAIGETLYLHQPWPSLKTWLSLSTILGGSV 834
I + + V + +SA P F + +L + +P + +LSL I+GG
Sbjct: 168 HTIGHVAATVSMSKVAVSFTHIIKSAEPAFSVLVSRFFLGETFP-IPVYLSLLPIIGGCA 226
Query: 833 IYVFTDNQFTVTAYTWAVAYLASMSIDFVYIKHVVMTIGLNTWGLVLYNNXXXXXXXXXX 654
+ T+ F + + A+ + ++ K + ++ +
Sbjct: 227 LAAVTELNFNMVGFMGAMISNLAFVFRNIFSKRGMKGKSVSGMNYYACLSIMSLVILTPF 286
Query: 653 XLVMGEFDQMKVDSSKVSNWLSFDVVLPVAXXXXXXXXXXXXXXXCRRAISATGFTVLGI 474
+ M K + +VV +A IS F++
Sbjct: 287 AIAMEGPQMWAAGWQKALAEVGPNVVWWIAAQSVFYHLYNQVSYMSLDQISPLTFSIGNT 346
Query: 473 VNKLLTVVINLLIWDKHASFVGTIGLLICMSGGVLYQQS 357
+ ++ +V +++I+ V +G I + G LY Q+
Sbjct: 347 MKRISVIVSSIIIFHTPVRAVNALGAAIAILGTFLYSQA 385
>sptr|Q94JS6|Q94JS6 Glucose-6-phosphate/phosphate translocator.
Length = 387
Score = 45.8 bits (107), Expect = 0.002
Identities = 56/279 (20%), Positives = 102/279 (36%), Gaps = 1/279 (0%)
Frame = -1
Query: 1190 LLSIINKWAIMKFPYPGALTALQYFTSVAGVLLCGQLKLIEHDGLNLRTMWKFL-PAAVM 1014
+ +I NK + FPYP + L A +L+ +L+E +L WK L P AV
Sbjct: 109 IFNIYNKKVLNAFPYPWLTSTLSLACGSAMMLVSWATRLVEAPKTDL-DFWKVLFPVAVA 167
Query: 1013 FYISIFTNSELLLHADVDTFIVFRSAVPIFVAIGETLYLHQPWPSLKTWLSLSTILGGSV 834
I + + V + +SA P F + L + +P + +LSL I+GG
Sbjct: 168 HTIGHVAATVSMSKVAVSFTHIIKSAEPAFSVLVSRFLLGETFP-VPVYLSLLPIIGGCG 226
Query: 833 IYVFTDNQFTVTAYTWAVAYLASMSIDFVYIKHVVMTIGLNTWGLVLYNNXXXXXXXXXX 654
+ T+ F + + A+ + ++ K + ++ +
Sbjct: 227 LAAVTELNFNMVGFMGAMISNLAFVFRNIFSKRGMKGKSVSGMNYYACLSIMSLVILTPF 286
Query: 653 XLVMGEFDQMKVDSSKVSNWLSFDVVLPVAXXXXXXXXXXXXXXXCRRAISATGFTVLGI 474
+ M K + DVV VA IS F++
Sbjct: 287 AIAMEGPQMWAAGWQKALAEVGPDVVWWVAAQSVFYHLYNQVSYMSLDEISPLTFSIGNT 346
Query: 473 VNKLLTVVINLLIWDKHASFVGTIGLLICMSGGVLYQQS 357
+ ++ +V +++I+ V +G I + G LY Q+
Sbjct: 347 MKRISVIVSSIIIFHTPVRPVNALGAAIAILGTFLYSQA 385
>sw|Q9NTN3|S35D_HUMAN UDP-glucuronic acid/UDP-N-acetylgalactosamine
transporter (UDP- GlcA/UDP-GalNAc transporter) (Solute
carrier family 35 member D1).
Length = 355
Score = 45.8 bits (107), Expect = 0.002
Identities = 55/293 (18%), Positives = 119/293 (40%), Gaps = 4/293 (1%)
Frame = -1
Query: 1220 VAAG-YCLSASLLSIINKWAIMKFPYPGAL-TALQYFTSVAGVLLCGQ-LKLIEHDGLNL 1050
+AAG Y +S+ L+ ++NK + + +P +L L + VL G+ L++++ L+
Sbjct: 44 LAAGFYGVSSFLIVVVNKSVLTNYRFPSSLCVGLGQMVATVAVLWVGKALRVVKFPDLDR 103
Query: 1049 RTMWKFLPAAVMFYISIFTNSELLLHADVDTFIVFRSAVPIFVAIGETLYLHQPWPSLKT 870
K P ++++ + T ++ F V R +F E + L + + S
Sbjct: 104 NVPRKTFPLPLLYFGNQITGLFSTKKLNLPMFTVLRRFSILFTMFAEGVLLKKTF-SWGI 162
Query: 869 WLSLSTILGGSVIYVFTDNQFTVTAYTWAVAYLASMSIDFVYIKHVVMTIGLNTWGLVLY 690
+++ ++ G+ + +D F + Y + + + + Y+K + + L +GL+ Y
Sbjct: 163 KMTVFAMIIGAFVAASSDLAFDLEGYAFILINDVLTAANGAYVKQKLDSKELGKYGLLYY 222
Query: 689 NNXXXXXXXXXXXLVMGEFDQMKVDSSKVSNWLSFDVVLPVAXXXXXXXXXXXXXXXCRR 510
N G+ + + + W +L C +
Sbjct: 223 NALFMILPTLAIAYFTGDAQK----AVEFEGWADTLFLLQFTLSCVMGFILMYATVLCTQ 278
Query: 509 AISATGFTVLGIVNKLLTVVINLLI-WDKHASFVGTIGLLICMSGGVLYQQST 354
SA T++G + +L I ++ D ++ IGL I ++G ++Y T
Sbjct: 279 YNSALTTTIVGCIKNILITYIGMVFGGDYIFTWTNFIGLNISIAGSLVYSYIT 331
>sptrnew|BAC99016|BAC99016 Nucleotide sugar transporter UGTrel8.
Length = 337
Score = 45.4 bits (106), Expect = 0.002
Identities = 56/284 (19%), Positives = 109/284 (38%), Gaps = 3/284 (1%)
Frame = -1
Query: 1208 YCLSASLLSIINKWAIMKFPYPGAL-TALQYFTSVAGVLLCGQL-KLIEHDGLNLRTMWK 1035
Y + L+ ++NK + + +P + + + +L +L K+I + + K
Sbjct: 34 YGTCSFLIVLVNKALLTTYGFPSPIFLGIGQMAATIMILYVSKLNKIIHFPDFDKKIPVK 93
Query: 1034 FLPAAVMFYISIFTNSELLLHADVDTFIVFRSAVPIFVAIGETLYLHQPWPSLKTWLSLS 855
P +++ + + + F V R + ET+ L + + SL LS+
Sbjct: 94 LFPLPLLYVGNHISGLSSTSKLSLPMFTVLRKFTIPLTLLLETIILGKQY-SLNIILSVF 152
Query: 854 TILGGSVIYVFTDNQFTVTAYTWAVAYLASMSIDFVYIKHVVMTIGLNTWGLVLYNNXXX 675
I+ G+ I +D F + Y + S + VY K + L +G++ YN
Sbjct: 153 AIILGAFIAAGSDLAFNLEGYIFVFLNDIFTSANGVYTKQKMDPKELGKYGVLFYNACFM 212
Query: 674 XXXXXXXXLVMGEFDQMKVDSSKVSNWLSFDVVLPVAXXXXXXXXXXXXXXXCRRAISAT 495
+ G+ Q +++ + W + +L C SA
Sbjct: 213 IIPTLIISVSTGDLQQ----ATEFNQWKNVVFILQFLLSCFLGFLLMYSTVLCSYYNSAL 268
Query: 494 GFTVLGIVNKLLTVVINLLIWDKHA-SFVGTIGLLICMSGGVLY 366
V+G + + I +LI + S + +GL ICM+GG+ Y
Sbjct: 269 TTAVVGAIKNVSVAYIGILIGGDYIFSLLNFVGLNICMAGGLRY 312
Database: /db/uniprot/tmp/swall
Posted date: Mar 5, 2004 7:45 PM
Number of letters in database: 442,889,342
Number of sequences in database: 1,395,590
Lambda K H
0.318 0.135 0.401
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 1,228,623,058
Number of Sequences: 1395590
Number of extensions: 26809946
Number of successful extensions: 74103
Number of sequences better than 10.0: 30
Number of HSP's better than 10.0 without gapping: 68891
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 73995
length of database: 442,889,342
effective HSP length: 127
effective length of database: 265,649,412
effective search space used: 97493334204
frameshift window, decay const: 50, 0.1
T: 12
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)

