BLASTX 2.2.6 [Apr-09-2003]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= 3802485.2.1
(1118 letters)
Database: /db/trembl-ebi/tmp/swall
1,272,877 sequences; 406,886,216 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
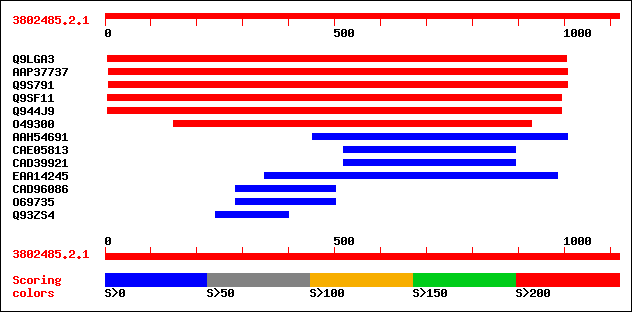
sptr|Q9LGA3|Q9LGA3 ESTs D22655(C0749). 557 e-157
sptrnew|AAP37737|AAP37737 At1g70770. 383 e-105
sptr|Q9S791|Q9S791 Hypothetical protein. 383 e-105
sptr|Q9SF11|Q9SF11 F26K24.17 protein. 251 1e-65
sptr|Q944J9|Q944J9 AT3g11880/F26K24_17. 251 1e-65
sptr|O49300|O49300 T26J12.6 protein. 245 7e-64
sptrnew|AAH54691|AAH54691 Hypothetical protein (Fragment). 43 0.007
sptrnew|CAE05813|CAE05813 OSJNBa0028M15.5 protein. 37 0.67
sptrnew|CAD39921|CAD39921 OSJNBa0065B15.26 protein. 37 0.67
sptrnew|EAA14245|EAA14245 ENSANGP00000015679 (Fragment). 36 1.1
sptrnew|CAD96086|CAD96086 POSSIBLE CONSERVED MEMBRANE PROTEIN. 33 7.4
sptr|O69735|O69735 Hypothetical 80.9 kDa protein (FtsK/spoIIIE f... 33 7.4
sptr|Q93ZS4|Q93ZS4 Putative serine/threonine protein kinase (Put... 33 7.4
>sptr|Q9LGA3|Q9LGA3 ESTs D22655(C0749).
Length = 586
Score = 557 bits (1435), Expect = e-157
Identities = 276/333 (82%), Positives = 298/333 (89%)
Frame = +2
Query: 5 ISSSYENQQDIQLMRFADYFGRAFVAVSASQFAWAKMFKESTVSKMVDIPLCHIPEAVIK 184
IS SYENQQDIQLMRFADYFGR+F +VSA+QF WAKMFKES VSKMVDIPLCHIPE V
Sbjct: 154 ISESYENQQDIQLMRFADYFGRSFASVSAAQFPWAKMFKESLVSKMVDIPLCHIPEPVRN 213
Query: 185 TASDWISQRSSDALGDFVLWCIDSIMSELSGPSAGPKGSKKVAQQSPRAQVAIFVVLAMT 364
TASDWI+QRS DALGDFV+WCIDSIMSELSG + G KGSKK AQQ+PRAQVAIFVVLA+T
Sbjct: 214 TASDWINQRSPDALGDFVMWCIDSIMSELSGQAVGAKGSKKAAQQTPRAQVAIFVVLALT 273
Query: 365 LRRKPDVLVNVMPKIMGNNKYLGQEKLPIIVWVIAQASQGDLVSGMFCWAHSLFPTLCAK 544
+RRKP+VL NV+PKIMGNNKYLGQEKLPIIVWVIAQASQGDLV+GMFCWAH LFPTLCAK
Sbjct: 274 VRRKPEVLTNVLPKIMGNNKYLGQEKLPIIVWVIAQASQGDLVTGMFCWAHFLFPTLCAK 333
Query: 545 SSGNPLARDLVLQLLERILSVTKARSILLNGAVRKGERLVPPVSFDLFMRATFPVSNARV 724
SGNP RDLVLQLLERILS KAR ILLNGAVRKGERL+PPV+FDLFMRA FPVS+ARV
Sbjct: 334 PSGNPQTRDLVLQLLERILSAPKARGILLNGAVRKGERLIPPVTFDLFMRAAFPVSSARV 393
Query: 725 KATERFEAAYPMIKELALAGPPGSKTVKQASQQLLPLCAKAMQENNAELTREAVDVFIWC 904
KATERFEAAYP IKELALAGPPGSKTVKQA+QQLLPLC KAMQENNA+LT E+ VFIWC
Sbjct: 394 KATERFEAAYPTIKELALAGPPGSKTVKQAAQQLLPLCVKAMQENNADLTGESAGVFIWC 453
Query: 905 LTQNAESYKXXERIYLENIEAXXAVLSKVVIDW 1003
LTQNAESYK ER++ EN+EA VLS +V W
Sbjct: 454 LTQNAESYKLWERLHPENVEASVVVLSTIVTKW 486
>sptrnew|AAP37737|AAP37737 At1g70770.
Length = 610
Score = 383 bits (983), Expect = e-105
Identities = 188/334 (56%), Positives = 246/334 (73%), Gaps = 1/334 (0%)
Frame = +2
Query: 8 SSSYENQQDIQLMRFADYFGRAFVAVSASQFAWAKMFKESTVSKMVDIPLCHIPEAVIKT 187
S SY +Q +IQLMRFADYFGRA VS+ QF W KMFKES +SK++++PL HIPE V KT
Sbjct: 159 SESYASQPEIQLMRFADYFGRALSGVSSVQFPWVKMFKESPLSKLIEVPLAHIPEPVYKT 218
Query: 188 ASDWISQRSSDALGDFVLWCIDSIMSELSGPSAGPKGSKKVAQQSP-RAQVAIFVVLAMT 364
+ DWI+ R +ALG FVLW D I+++L+ G KG KK QQ+ ++QVAIFV LAM
Sbjct: 219 SVDWINHRPIEALGAFVLWAFDCILTDLAAQQGGAKGGKKGGQQTTSKSQVAIFVALAMV 278
Query: 365 LRRKPDVLVNVMPKIMGNNKYLGQEKLPIIVWVIAQASQGDLVSGMFCWAHSLFPTLCAK 544
LRRKPD L NV+P + N KY GQ+KLP+ VW++AQASQGD+ G++ WAH+L P + K
Sbjct: 279 LRRKPDALTNVLPTLRENPKYQGQDKLPVTVWMMAQASQGDIAVGLYSWAHNLLPVVGNK 338
Query: 545 SSGNPLARDLVLQLLERILSVTKARSILLNGAVRKGERLVPPVSFDLFMRATFPVSNARV 724
+ NP +RDL+LQL+E+IL+ KAR+IL+NGAVRKGERL+PP SF++ +R TFP S+ARV
Sbjct: 339 NC-NPQSRDLILQLVEKILTNPKARTILVNGAVRKGERLIPPPSFEILLRLTFPASSARV 397
Query: 725 KATERFEAAYPMIKELALAGPPGSKTVKQASQQLLPLCAKAMQENNAELTREAVDVFIWC 904
KATERFEA YP++KE+ALAG PGSK +KQ +QQ+ K E N L +EA + IW
Sbjct: 398 KATERFEAIYPLLKEVALAGAPGSKAMKQVTQQIFTFALKLAGEGNPVLAKEATAIAIWS 457
Query: 905 LTQNAESYKXXERIYLENIEAXXAVLSKVVIDWR 1006
+TQN + K + +Y EN+EA AVL K+V +W+
Sbjct: 458 VTQNFDCCKHWDNLYKENLEASVAVLKKLVEEWK 491
>sptr|Q9S791|Q9S791 Hypothetical protein.
Length = 610
Score = 383 bits (983), Expect = e-105
Identities = 188/334 (56%), Positives = 246/334 (73%), Gaps = 1/334 (0%)
Frame = +2
Query: 8 SSSYENQQDIQLMRFADYFGRAFVAVSASQFAWAKMFKESTVSKMVDIPLCHIPEAVIKT 187
S SY +Q +IQLMRFADYFGRA VS+ QF W KMFKES +SK++++PL HIPE V KT
Sbjct: 159 SESYASQPEIQLMRFADYFGRALSGVSSVQFPWVKMFKESPLSKLIEVPLAHIPEPVYKT 218
Query: 188 ASDWISQRSSDALGDFVLWCIDSIMSELSGPSAGPKGSKKVAQQSP-RAQVAIFVVLAMT 364
+ DWI+ R +ALG FVLW D I+++L+ G KG KK QQ+ ++QVAIFV LAM
Sbjct: 219 SVDWINHRPIEALGAFVLWAFDCILTDLAAQQGGAKGGKKGGQQTTSKSQVAIFVALAMV 278
Query: 365 LRRKPDVLVNVMPKIMGNNKYLGQEKLPIIVWVIAQASQGDLVSGMFCWAHSLFPTLCAK 544
LRRKPD L NV+P + N KY GQ+KLP+ VW++AQASQGD+ G++ WAH+L P + K
Sbjct: 279 LRRKPDALTNVLPTLRENPKYQGQDKLPVTVWMMAQASQGDIAVGLYSWAHNLLPVVGNK 338
Query: 545 SSGNPLARDLVLQLLERILSVTKARSILLNGAVRKGERLVPPVSFDLFMRATFPVSNARV 724
+ NP +RDL+LQL+E+IL+ KAR+IL+NGAVRKGERL+PP SF++ +R TFP S+ARV
Sbjct: 339 NC-NPQSRDLILQLVEKILTNPKARTILVNGAVRKGERLIPPPSFEILLRLTFPASSARV 397
Query: 725 KATERFEAAYPMIKELALAGPPGSKTVKQASQQLLPLCAKAMQENNAELTREAVDVFIWC 904
KATERFEA YP++KE+ALAG PGSK +KQ +QQ+ K E N L +EA + IW
Sbjct: 398 KATERFEAIYPLLKEVALAGAPGSKAMKQVTQQIFTFALKLAGEGNPVLAKEATAIAIWS 457
Query: 905 LTQNAESYKXXERIYLENIEAXXAVLSKVVIDWR 1006
+TQN + K + +Y EN+EA AVL K+V +W+
Sbjct: 458 VTQNFDCCKHWDNLYKENLEASVAVLKKLVEEWK 491
>sptr|Q9SF11|Q9SF11 F26K24.17 protein.
Length = 459
Score = 251 bits (642), Expect = 1e-65
Identities = 143/338 (42%), Positives = 208/338 (61%), Gaps = 8/338 (2%)
Frame = +2
Query: 5 ISSSYENQQDIQLMRFADYFGRAFVAVSASQFAWAKMFKESTVSK---MVDIPLCHIPEA 175
IS S+ + QL++F DY + +S+ Q+ W MFK S K M+D+PL HIP +
Sbjct: 43 ISKSHAFVPEEQLLKFVDYLE---IKLSSVQYLWLDMFKGSPCPKLIDMIDVPLSHIPVS 99
Query: 176 VIKTASDWISQRSSDALGDFVLWCIDSIMSELSGPSAGPKGSKKVAQQSPRAQVAIFVVL 355
V T+ +W+ + S L FV+W ++ +++ L P G G ++ + + + VA+FV L
Sbjct: 100 VYDTSVEWLDKFSIGLLCAFVVWSLNRLLTILEPPQQG--GHQR--RTTSKFHVAVFVAL 155
Query: 356 AMTLRRKPDVLVNVMPKIMGNNKYLGQEKLPIIVWVIAQASQGDLVSGMFCWAHSLFPT- 532
AM LR +P+ LV V+P + ++Y G +KLPI+VW++AQASQGDL G++ W+ +L P
Sbjct: 156 AMVLRNEPNTLVIVLPTLK-EDEYQGHDKLPILVWMMAQASQGDLSVGLYSWSCNLLPVF 214
Query: 533 ----LCAKSSGNPLARDLVLQLLERILSVTKARSILLNGAVRKGERLVPPVSFDLFMRAT 700
L S N + DL+LQL E ILS AR+IL+NG V +RL+ P +F+L MR T
Sbjct: 215 YQENLLPVSRSNSQSMDLILQLAEMILSNLDARTILVNGTVIDKQRLISPYAFELLMRLT 274
Query: 701 FPVSNARVKATERFEAAYPMIKELALAGPPGSKTVKQASQQLLPLCAKAMQENNAELTRE 880
FP S+ RVKATERFEA YP++KE+ALA PGS+ +KQ +QQ+ N L +E
Sbjct: 275 FPASSERVKATERFEAIYPLLKEVALACEPGSELMKQVTQQIFHYSLIIAGRRNLVLAKE 334
Query: 881 AVDVFIWCLTQNAESYKXXERIYLENIEAXXAVLSKVV 994
A + +W LT+N + K E++Y EN EA AVL K+V
Sbjct: 335 ATAIAVWSLTENVDCCKQWEKLYWENKEASVAVLKKLV 372
>sptr|Q944J9|Q944J9 AT3g11880/F26K24_17.
Length = 443
Score = 251 bits (642), Expect = 1e-65
Identities = 143/338 (42%), Positives = 208/338 (61%), Gaps = 8/338 (2%)
Frame = +2
Query: 5 ISSSYENQQDIQLMRFADYFGRAFVAVSASQFAWAKMFKESTVSK---MVDIPLCHIPEA 175
IS S+ + QL++F DY + +S+ Q+ W MFK S K M+D+PL HIP +
Sbjct: 27 ISKSHAFVPEEQLLKFVDYLE---IKLSSVQYLWLDMFKGSPCPKLIDMIDVPLSHIPVS 83
Query: 176 VIKTASDWISQRSSDALGDFVLWCIDSIMSELSGPSAGPKGSKKVAQQSPRAQVAIFVVL 355
V T+ +W+ + S L FV+W ++ +++ L P G G ++ + + + VA+FV L
Sbjct: 84 VYDTSVEWLDKFSIGLLCAFVVWSLNRLLTILEPPQQG--GHQR--RTTSKFHVAVFVAL 139
Query: 356 AMTLRRKPDVLVNVMPKIMGNNKYLGQEKLPIIVWVIAQASQGDLVSGMFCWAHSLFPT- 532
AM LR +P+ LV V+P + ++Y G +KLPI+VW++AQASQGDL G++ W+ +L P
Sbjct: 140 AMVLRNEPNTLVIVLPTLK-EDEYQGHDKLPILVWMMAQASQGDLSVGLYSWSCNLLPVF 198
Query: 533 ----LCAKSSGNPLARDLVLQLLERILSVTKARSILLNGAVRKGERLVPPVSFDLFMRAT 700
L S N + DL+LQL E ILS AR+IL+NG V +RL+ P +F+L MR T
Sbjct: 199 YQENLLPVSRSNSQSMDLILQLAEMILSNLDARTILVNGTVIDKQRLISPYAFELLMRLT 258
Query: 701 FPVSNARVKATERFEAAYPMIKELALAGPPGSKTVKQASQQLLPLCAKAMQENNAELTRE 880
FP S+ RVKATERFEA YP++KE+ALA PGS+ +KQ +QQ+ N L +E
Sbjct: 259 FPASSERVKATERFEAIYPLLKEVALACEPGSELMKQVTQQIFHYSLIIAGRRNLVLAKE 318
Query: 881 AVDVFIWCLTQNAESYKXXERIYLENIEAXXAVLSKVV 994
A + +W LT+N + K E++Y EN EA AVL K+V
Sbjct: 319 ATAIAVWSLTENVDCCKQWEKLYWENKEASVAVLKKLV 356
>sptr|O49300|O49300 T26J12.6 protein.
Length = 1299
Score = 245 bits (626), Expect = 7e-64
Identities = 131/264 (49%), Positives = 182/264 (68%), Gaps = 4/264 (1%)
Frame = +2
Query: 149 IPLCHIPEAVIKTASDWISQRSSDALGDFVLWCIDSIMSELSGPSAGPKGSKKVAQQ-SP 325
IPL HIPEAV KT++DWI+QR +ALG FVLW +D I+++L+ G KG KK AQQ S
Sbjct: 170 IPLSHIPEAVYKTSADWINQRPIEALGAFVLWGLDCILADLAVQQGGVKGGKKGAQQASS 229
Query: 326 RAQVAIFVVLAMTLRRKPDVLVNVMPKIMGNNKYLGQEKLPIIVWVIAQASQGDLVSGMF 505
++QVAIFV +AM LR+KPD L N++P + N KY GQ+KLP+ VW++AQASQGD+ G++
Sbjct: 230 KSQVAIFVAVAMVLRKKPDALTNILPTLRENPKYQGQDKLPVTVWMMAQASQGDISVGLY 289
Query: 506 CWAHSLFPTLCAKSSGNPLARDLVLQLLERILSVTKARSILLNGAVRKGERLVPPVSFDL 685
WAH+L P + +KS NP +RDL+LQL+ERILS KAR+IL+NGAVRKGERL+PP SF++
Sbjct: 290 SWAHNLLPVVSSKSC-NPQSRDLILQLVERILSNPKARTILVNGAVRKGERLIPPPSFEI 348
Query: 686 FMRATFPVSNARVKA---TERFEAAYPMIKELALAGPPGSKTVKQASQQLLPLCAKAMQE 856
+R TFP S+ARVK T+ +A+ ++K+L S + A L K++++
Sbjct: 349 LVRLTFPASSARVKENLYTDNLKASVAVLKKLIGEWKERSVKLTPAETLTLNQTMKSLRQ 408
Query: 857 NNAELTREAVDVFIWCLTQNAESY 928
N E E + L ++A+ Y
Sbjct: 409 KNEEALTEGGNGVSQSLYKDADKY 432
>sptrnew|AAH54691|AAH54691 Hypothetical protein (Fragment).
Length = 304
Score = 43.1 bits (100), Expect = 0.007
Identities = 43/189 (22%), Positives = 79/189 (41%), Gaps = 4/189 (2%)
Frame = +2
Query: 452 IVWVIAQASQGDLVSGMFCWAHSLFPTLCAKSSGNPLARDLVLQLLERILSVTKARSILL 631
I+W + QA DL G+ W + P L K+ + LER+L+ L
Sbjct: 46 IMWALGQAGFYDLSQGIRVWLGIMLPVLGMKALSA-----YAIAYLERLLT--------L 92
Query: 632 NGAVRKGERLVPPVSFDLFMRATFPVSNARVKAT-ERFEAAYPMIKELALAGPPGSKT-- 802
+ + KG ++ P F + + NA ++ E+ YP +K LA P S
Sbjct: 93 HANLTKGFGIMGPKEFFPLLDFAYMPKNALSQSLQEQLCRLYPRLKVLAFGAKPESTLHT 152
Query: 803 -VKQASQQLLPLCAKAMQENNAELTREAVDVFIWCLTQNAESYKXXERIYLENIEAXXAV 979
+ P C AM++ EL R + CL+ +++S ++Y +++ +
Sbjct: 153 YFPPFLSRATPSCPGAMKK---ELLRSLTE----CLSVDSQSLSVWRQLYTKHLPQSSLL 205
Query: 980 LSKVVIDWR 1006
L+ ++ W+
Sbjct: 206 LNHLLKTWK 214
>sptrnew|CAE05813|CAE05813 OSJNBa0028M15.5 protein.
Length = 1055
Score = 36.6 bits (83), Expect = 0.67
Identities = 33/134 (24%), Positives = 58/134 (43%), Gaps = 9/134 (6%)
Frame = +2
Query: 518 SLFPTLCAKSSGNPLARDLVLQLLERILSVTKARSILLNGAVRKGERLVPPVSFDLFMRA 697
+ F LC+K+ L +D+++Q+ E I+ + + K E++ PP FD+ M
Sbjct: 629 NFFKRLCSKT----LNKDVLVQMNEEIIVL-----------LCKLEKIFPPALFDVMMHL 673
Query: 698 TFP-VSNARVKATERFEAAYPMIKELAL--------AGPPGSKTVKQASQQLLPLCAKAM 850
V A ++ ++ YP+ + L A P GS + + L C+K M
Sbjct: 674 PVHLVEEALLRGPVQYGWMYPIERRLYTLKRYVRNGARPEGSIAEAYIADECLTFCSKYM 733
Query: 851 QENNAELTREAVDV 892
+ REA +V
Sbjct: 734 DDVETRFNREARNV 747
>sptrnew|CAD39921|CAD39921 OSJNBa0065B15.26 protein.
Length = 1055
Score = 36.6 bits (83), Expect = 0.67
Identities = 33/134 (24%), Positives = 58/134 (43%), Gaps = 9/134 (6%)
Frame = +2
Query: 518 SLFPTLCAKSSGNPLARDLVLQLLERILSVTKARSILLNGAVRKGERLVPPVSFDLFMRA 697
+ F LC+K+ L +D+++Q+ E I+ + + K E++ PP FD+ M
Sbjct: 629 NFFKRLCSKT----LNKDVLVQMNEEIIVL-----------LCKLEKIFPPALFDVMMHL 673
Query: 698 TFP-VSNARVKATERFEAAYPMIKELAL--------AGPPGSKTVKQASQQLLPLCAKAM 850
V A ++ ++ YP+ + L A P GS + + L C+K M
Sbjct: 674 PVHLVEEALLRGPVQYGWMYPIERRLYTLKRYVRNGARPEGSIAEAYIADECLTFCSKYM 733
Query: 851 QENNAELTREAVDV 892
+ REA +V
Sbjct: 734 DDVETRFNREARNV 747
>sptrnew|EAA14245|EAA14245 ENSANGP00000015679 (Fragment).
Length = 433
Score = 35.8 bits (81), Expect = 1.1
Identities = 45/217 (20%), Positives = 82/217 (37%), Gaps = 4/217 (1%)
Frame = +2
Query: 347 VVLAMTLRRKPDVLVNVMPK-IMGNNKYLGQEKLPI-IVWVIAQASQGDLVSGMFCWAHS 520
V+L P VN + + + N Y + + + ++W + Q DL G+ W
Sbjct: 194 VILQAIAMHYPSACVNNLARNAILRNSYQNRHNIGLSLLWALGQGGYNDLDVGLKVWQDI 253
Query: 521 LFPTLCAKSSGNPLARDLVLQLLERILSVTKARSILLNGAVRKGERLVPPVSFDLFMRAT 700
+ P + K+ N D V+ RIL + +A + L G+ F+
Sbjct: 254 MVPVMELKNY-NRFTSDYVV----RILRLHRAHRLTLGGSE--------------FLTIL 294
Query: 701 FPVSNARVKATERFEAAYPMIKELALAGPPGSKTVKQASQQLLPLCAKAMQENNAELTRE 880
++ E EAA +++ + P S T + +N + +TR
Sbjct: 295 SSLTTQPKACRELDEAAQLLVERYVFSAPKASATFTM------------LFKNVSFITRP 342
Query: 881 AVDVF--IWCLTQNAESYKXXERIYLENIEAXXAVLS 985
+ + CL ++ ES +Y N+E A+LS
Sbjct: 343 EMIYYGLALCLLEDPESAAVWLGLYRSNVETSLAILS 379
>sptrnew|CAD96086|CAD96086 POSSIBLE CONSERVED MEMBRANE PROTEIN.
Length = 747
Score = 33.1 bits (74), Expect = 7.4
Identities = 21/74 (28%), Positives = 36/74 (48%), Gaps = 1/74 (1%)
Frame = +2
Query: 284 AGPKGSKKVAQQSPRAQVAIFVVLAMTLRRKPDVLVNVMPKIMGNNKYLGQEKLPIIVWV 463
AGP G S +++ ++L++ PD + ++ G + +LG EKLP V
Sbjct: 471 AGPHGMLIGTTGSGKSEFLRTLILSLVAMTHPDQVNLLLTDFKGGSTFLGMEKLPHTAAV 530
Query: 464 IA-QASQGDLVSGM 502
+ A + +LVS M
Sbjct: 531 VTNMAEEAELVSRM 544
>sptr|O69735|O69735 Hypothetical 80.9 kDa protein (FtsK/spoIIIE
family protein).
Length = 747
Score = 33.1 bits (74), Expect = 7.4
Identities = 21/74 (28%), Positives = 36/74 (48%), Gaps = 1/74 (1%)
Frame = +2
Query: 284 AGPKGSKKVAQQSPRAQVAIFVVLAMTLRRKPDVLVNVMPKIMGNNKYLGQEKLPIIVWV 463
AGP G S +++ ++L++ PD + ++ G + +LG EKLP V
Sbjct: 471 AGPHGMLIGTTGSGKSEFLRTLILSLVAMTHPDQVNLLLTDFKGGSTFLGMEKLPHTAAV 530
Query: 464 IA-QASQGDLVSGM 502
+ A + +LVS M
Sbjct: 531 VTNMAEEAELVSRM 544
>sptr|Q93ZS4|Q93ZS4 Putative serine/threonine protein kinase
(Putative receptor kinase).
Length = 632
Score = 33.1 bits (74), Expect = 7.4
Identities = 24/60 (40%), Positives = 30/60 (50%), Gaps = 7/60 (11%)
Frame = -3
Query: 399 ITFTSTSGFLLKVMARTTKIATWAL------GDCCATFFEPLG-PADGPDNSDMMLSIHH 241
I++ + SG L KV ART K+ AL C+ EPL P DGPD S + HH
Sbjct: 177 ISYNNLSGSLPKVSARTFKVIGNALICGPKAVSNCSAVPEPLTLPQDGPDESGTRTNGHH 236
Database: /db/trembl-ebi/tmp/swall
Posted date: Sep 26, 2003 8:29 PM
Number of letters in database: 406,886,216
Number of sequences in database: 1,272,877
Lambda K H
0.318 0.135 0.401
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 769,754,445
Number of Sequences: 1272877
Number of extensions: 16797843
Number of successful extensions: 45776
Number of sequences better than 10.0: 13
Number of HSP's better than 10.0 without gapping: 43891
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 45752
length of database: 406,886,216
effective HSP length: 124
effective length of database: 249,049,468
effective search space used: 61764268064
frameshift window, decay const: 50, 0.1
T: 12
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)

