BLASTX 2.2.6 [Apr-09-2003]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= 3696537.2.1
(592 letters)
Database: /db/trembl-ebi/tmp/swall
1,272,877 sequences; 406,886,216 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
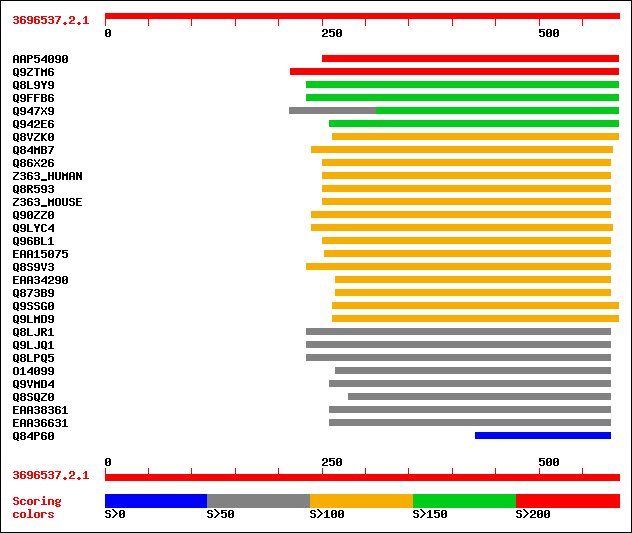
sptrnew|AAP54090|AAP54090 Putative PGPD14 protein (Pollen germin... 213 2e-54
sptr|Q9ZTM6|Q9ZTM6 PGPD14. 201 5e-51
sptr|Q8L9Y9|Q8L9Y9 PGPD14 protein. 184 9e-46
sptr|Q9FFB6|Q9FFB6 PGPD14 protein (AT5g22920/MRN17_15). 183 1e-45
sptr|Q947X9|Q947X9 Hypothetical 39.6 kDa protein. 157 1e-37
sptr|Q942E6|Q942E6 Putative PGPD14 protein (Pollen germination r... 152 2e-36
sptr|Q8VZK0|Q8VZK0 Hypothetical protein. 147 1e-34
sptr|Q84MB7|Q84MB7 At3g62970. 127 7e-29
sptr|Q86X26|Q86X26 Similar to zinc finger protein 363. 117 1e-25
sw|Q96PM5|Z363_HUMAN Zinc finger protein 363 (CH-rich interactin... 117 1e-25
sptr|Q8R593|Q8R593 RIKEN cDNA 6720407C15 gene. 116 2e-25
sw|Q9CR50|Z363_MOUSE Zinc finger protein 363 (CH-rich interactin... 116 2e-25
sptr|Q90ZZ0|Q90ZZ0 Zinc finger protein. 115 3e-25
sptr|Q9LYC4|Q9LYC4 Hypothetical 30.9 kDa protein. 115 5e-25
sptr|Q96BL1|Q96BL1 Hypothetical protein (Fragment). 108 6e-23
sptrnew|EAA15075|EAA15075 ENSANGP00000012255 (Fragment). 103 1e-21
sptr|Q8S9V3|Q8S9V3 Putative zinc finger protein. 102 4e-21
sptrnew|EAA34290|EAA34290 Hypothetical protein. 101 6e-21
sptr|Q873B9|Q873B9 Hypothetical protein B24N11.050. 101 6e-21
sptr|Q9SSG0|Q9SSG0 F25A4.27 protein. 101 7e-21
sptr|Q9LMD9|Q9LMD9 F14D16.3. 101 7e-21
sptr|Q8LJR1|Q8LJR1 Putative zinc finger protein. 100 1e-20
sptr|Q9LJQ1|Q9LJQ1 Gb|AAD55299.1. 99 3e-20
sptr|Q8LPQ5|Q8LPQ5 AT3g18290/MIE15_8. 99 3e-20
sptr|O14099|O14099 Zinc finger protein. 96 2e-19
sptr|Q9VMD4|Q9VMD4 CG16947 protein. 94 2e-18
sptr|Q8SQZ0|Q8SQZ0 LIM domain-containing protein. 57 2e-07
sptrnew|EAA38361|EAA38361 GLP_375_16333_15149. 51 1e-05
sptrnew|EAA36631|EAA36631 GLP_115_10715_10044. 51 1e-05
sptr|Q84P60|Q84P60 Putative zinc finger protein (Fragment). 50 3e-05
>sptrnew|AAP54090|AAP54090 Putative PGPD14 protein (Pollen
germination related protein).
Length = 266
Score = 213 bits (541), Expect = 2e-54
Identities = 91/114 (79%), Positives = 100/114 (87%)
Frame = -2
Query: 591 MHHNCPVCMEYLFDSMKAISVLHCGHTIHLECLYEMRAHQQFSCPVCLRSACNMSDIWQK 412
MHHNCPVC EYLFDS K ISVLHCGHTIHLECL MRAH F+CPVC RSAC+MSD W+K
Sbjct: 142 MHHNCPVCFEYLFDSTKDISVLHCGHTIHLECLNVMRAHHHFACPVCSRSACDMSDAWKK 201
Query: 411 LDQQVAASPMPAIYQKKMVWILCNDCGVTSNVQFHILAHKCPGCSSYNTRQTRG 250
LD++VAA+PMP YQKKM+WILCNDCG TSNV FH+LA KCPGCSSYNTR+TRG
Sbjct: 202 LDEEVAATPMPEFYQKKMIWILCNDCGATSNVNFHVLAQKCPGCSSYNTRETRG 255
>sptr|Q9ZTM6|Q9ZTM6 PGPD14.
Length = 285
Score = 201 bits (511), Expect = 5e-51
Identities = 85/126 (67%), Positives = 103/126 (81%)
Frame = -2
Query: 591 MHHNCPVCMEYLFDSMKAISVLHCGHTIHLECLYEMRAHQQFSCPVCLRSACNMSDIWQK 412
MHHNCPVC EY+FD+ K I+VL CGHT+HLEC+ +M H Q+SCPVC RS C+MS +W+K
Sbjct: 158 MHHNCPVCFEYVFDTTKNITVLPCGHTMHLECVMQMEQHNQYSCPVCSRSYCDMSRVWEK 217
Query: 411 LDQQVAASPMPAIYQKKMVWILCNDCGVTSNVQFHILAHKCPGCSSYNTRQTRGDPAACS 232
LDQ+VA++PMP +YQ KMVWILCNDCG TS V FHI+A KCP C SYNTRQTRG P++CS
Sbjct: 218 LDQEVASTPMPEMYQNKMVWILCNDCGETSEVNFHIVARKCPNCKSYNTRQTRGGPSSCS 277
Query: 231 RV*EQI 214
E+I
Sbjct: 278 SRIEEI 283
>sptr|Q8L9Y9|Q8L9Y9 PGPD14 protein.
Length = 274
Score = 184 bits (466), Expect = 9e-46
Identities = 76/122 (62%), Positives = 98/122 (80%), Gaps = 2/122 (1%)
Frame = -2
Query: 591 MHHNCPVCMEYLFDSMKAISVLHCGHTIHLECLYEMRAHQQFSCPVCLRSACNMSDIWQK 412
MHHNCPVC EYLFDS + I+VL CGHT+HLEC +M H +++CPVC +S C+MS++W+K
Sbjct: 142 MHHNCPVCFEYLFDSTRDITVLRCGHTMHLECTKDMGLHNRYTCPVCSKSICDMSNLWKK 201
Query: 411 LDQQVAASPMPAIYQKKMVWILCNDCGVTSNVQFHILAHKCPGCSSYNTRQTR--GDPAA 238
LD++VAA PMP +Y+ KMVWILCNDCG +NV+FH++AHKC C SYNTRQT+ D +
Sbjct: 202 LDEEVAAYPMPKLYENKMVWILCNDCGSNTNVRFHLIAHKCSSCGSYNTRQTQRGSDSHS 261
Query: 237 CS 232
CS
Sbjct: 262 CS 263
>sptr|Q9FFB6|Q9FFB6 PGPD14 protein (AT5g22920/MRN17_15).
Length = 291
Score = 183 bits (465), Expect = 1e-45
Identities = 76/122 (62%), Positives = 98/122 (80%), Gaps = 2/122 (1%)
Frame = -2
Query: 591 MHHNCPVCMEYLFDSMKAISVLHCGHTIHLECLYEMRAHQQFSCPVCLRSACNMSDIWQK 412
MHHNCPVC EYLFDS + I+VL CGHT+HLEC +M H +++CPVC +S C+MS++W+K
Sbjct: 159 MHHNCPVCFEYLFDSTRDITVLRCGHTMHLECTKDMGLHNRYTCPVCSKSICDMSNLWKK 218
Query: 411 LDQQVAASPMPAIYQKKMVWILCNDCGVTSNVQFHILAHKCPGCSSYNTRQTR--GDPAA 238
LD++VAA PMP +Y+ KMVWILCNDCG +NV+FH++AHKC C SYNTRQT+ D +
Sbjct: 219 LDEEVAAYPMPKMYENKMVWILCNDCGSNTNVRFHLIAHKCSSCGSYNTRQTQRGSDSHS 278
Query: 237 CS 232
CS
Sbjct: 279 CS 280
>sptr|Q947X9|Q947X9 Hypothetical 39.6 kDa protein.
Length = 349
Score = 157 bits (396), Expect = 1e-37
Identities = 73/93 (78%), Positives = 78/93 (83%)
Frame = -2
Query: 591 MHHNCPVCMEYLFDSMKAISVLHCGHTIHLECLYEMRAHQQFSCPVCLRSACNMSDIWQK 412
MHHNCPVC EYLFDS K IS LHCGHTIHLECLYEMR+HQQFSCPVCLRSAC+MS WQK
Sbjct: 208 MHHNCPVCFEYLFDSTKDISALHCGHTIHLECLYEMRSHQQFSCPVCLRSACDMSHAWQK 267
Query: 411 LDQQVAASPMPAIYQKKMVWILCNDCGVTSNVQ 313
LDQ+VAASPMP IYQKKMV C + S+VQ
Sbjct: 268 LDQEVAASPMPVIYQKKMV---CKFSYLCSSVQ 297
Score = 66.2 bits (160), Expect = 3e-10
Identities = 31/54 (57%), Positives = 41/54 (75%), Gaps = 1/54 (1%)
Frame = -1
Query: 370 SEEDGMDPLQRLWRDVQRAVPHLGAQVPWMQLLQHPADEGR-SSRML*SLRANK 212
S + MDP+QRLW D++RA+PH+G QVP MQLLQHPAD+ R R+L SL + +
Sbjct: 295 SVQPDMDPMQRLWDDIERAIPHIGTQVPRMQLLQHPADKSRPCCRVLQSLNSEE 348
>sptr|Q942E6|Q942E6 Putative PGPD14 protein (Pollen germination
related protein).
Length = 299
Score = 152 bits (385), Expect = 2e-36
Identities = 65/111 (58%), Positives = 83/111 (74%)
Frame = -2
Query: 591 MHHNCPVCMEYLFDSMKAISVLHCGHTIHLECLYEMRAHQQFSCPVCLRSACNMSDIWQK 412
MHH+CP+C EYLF+S +SVL CGHTIH++CL EM H QF+CP+C +S C+MS W++
Sbjct: 190 MHHDCPICFEYLFESTNDVSVLPCGHTIHVKCLREMEEHCQFACPLCSKSVCDMSKAWER 249
Query: 411 LDQQVAASPMPAIYQKKMVWILCNDCGVTSNVQFHILAHKCPGCSSYNTRQ 259
LD+++A + KMV ILCNDCG TS VQFH++AHKC C SYNTRQ
Sbjct: 250 LDEELAT--ISDTCDNKMVRILCNDCGATSEVQFHLIAHKCQKCKSYNTRQ 298
>sptr|Q8VZK0|Q8VZK0 Hypothetical protein.
Length = 267
Score = 147 bits (370), Expect = 1e-34
Identities = 60/110 (54%), Positives = 79/110 (71%)
Frame = -2
Query: 591 MHHNCPVCMEYLFDSMKAISVLHCGHTIHLECLYEMRAHQQFSCPVCLRSACNMSDIWQK 412
M H+CP+C EYLFDS+K +V+ CGHT+H+EC EM +F CP+C RS +MS WQ+
Sbjct: 149 MRHHCPICYEYLFDSLKDTNVMKCGHTMHVECYNEMIKRDKFCCPICSRSVIDMSKTWQR 208
Query: 411 LDQQVAASPMPAIYQKKMVWILCNDCGVTSNVQFHILAHKCPGCSSYNTR 262
LD+++ A+ MP+ Y+ K VWILCNDC T+ V FHI+ KC C SYNTR
Sbjct: 209 LDEEIEATAMPSDYRDKKVWILCNDCNDTTEVHFHIIGQKCGHCRSYNTR 258
>sptr|Q84MB7|Q84MB7 At3g62970.
Length = 276
Score = 127 bits (320), Expect = 7e-29
Identities = 59/118 (50%), Positives = 77/118 (65%), Gaps = 2/118 (1%)
Frame = -2
Query: 585 HNCPVCMEYLFDSMKAISVLHCGHTIHLECLYEMRAHQQFSCPVCLRSACNMSDIWQKLD 406
++CPVC EYLFDS+KA V+ CGHT+H++C +M Q+ CP+C +S +MS W LD
Sbjct: 157 NSCPVCYEYLFDSVKAAHVMKCGHTMHMDCFEQMINENQYRCPICAKSMVDMSPSWHLLD 216
Query: 405 QQVAASPMPAIYQKKMVWILCNDCGVTSNVQFHILAHKCPGCSSYNTRQ--TRGDPAA 238
+++A+ MP Y K V ILCNDC S FHIL HKC C SYNTR+ T DP +
Sbjct: 217 FEISATEMPVEY-KFEVSILCNDCNKGSKAMFHILGHKCSDCGSYNTRRISTPQDPVS 273
>sptr|Q86X26|Q86X26 Similar to zinc finger protein 363.
Length = 261
Score = 117 bits (292), Expect = 1e-25
Identities = 54/111 (48%), Positives = 69/111 (62%)
Frame = -2
Query: 582 NCPVCMEYLFDSMKAISVLHCGHTIHLECLYEMRAHQQFSCPVCLRSACNMSDIWQKLDQ 403
NCP+C+E + S VL CGH +H C YE + + CP+C+ SA +M+ W++LD
Sbjct: 144 NCPICLEDIHTSRVVAHVLPCGHLLHRTC-YEEMLKEGYRCPLCMHSALDMTRYWRQLDD 202
Query: 402 QVAASPMPAIYQKKMVWILCNDCGVTSNVQFHILAHKCPGCSSYNTRQTRG 250
+VA +PMP+ YQ V ILCNDC S VQFHIL KC C SYNT Q G
Sbjct: 203 EVAQTPMPSEYQNMTVDILCNDCNGRSTVQFHILGMKCKICESYNTAQAGG 253
>sw|Q96PM5|Z363_HUMAN Zinc finger protein 363 (CH-rich interacting
match with PLAG1) (Androgen receptor
N-terminal-interacting protein).
Length = 261
Score = 117 bits (292), Expect = 1e-25
Identities = 54/111 (48%), Positives = 69/111 (62%)
Frame = -2
Query: 582 NCPVCMEYLFDSMKAISVLHCGHTIHLECLYEMRAHQQFSCPVCLRSACNMSDIWQKLDQ 403
NCP+C+E + S VL CGH +H C YE + + CP+C+ SA +M+ W++LD
Sbjct: 144 NCPICLEDIHTSRVVAHVLPCGHLLHRTC-YEEMLKEGYRCPLCMHSALDMTRYWRQLDD 202
Query: 402 QVAASPMPAIYQKKMVWILCNDCGVTSNVQFHILAHKCPGCSSYNTRQTRG 250
+VA +PMP+ YQ V ILCNDC S VQFHIL KC C SYNT Q G
Sbjct: 203 EVAQTPMPSEYQNMTVDILCNDCNGRSTVQFHILGMKCKICESYNTAQAGG 253
>sptr|Q8R593|Q8R593 RIKEN cDNA 6720407C15 gene.
Length = 261
Score = 116 bits (290), Expect = 2e-25
Identities = 54/111 (48%), Positives = 69/111 (62%)
Frame = -2
Query: 582 NCPVCMEYLFDSMKAISVLHCGHTIHLECLYEMRAHQQFSCPVCLRSACNMSDIWQKLDQ 403
NCP+C+E + S VL CGH +H C YE + + CP+C+ SA +M+ W++LD
Sbjct: 144 NCPICLEDIHTSRVVAHVLPCGHLLHRTC-YEEMLKEGYRCPLCMHSALDMTRYWRQLDT 202
Query: 402 QVAASPMPAIYQKKMVWILCNDCGVTSNVQFHILAHKCPGCSSYNTRQTRG 250
+VA +PMP+ YQ V ILCNDC S VQFHIL KC C SYNT Q G
Sbjct: 203 EVAQTPMPSEYQNVTVDILCNDCNGRSTVQFHILGMKCKLCDSYNTAQAGG 253
>sw|Q9CR50|Z363_MOUSE Zinc finger protein 363 (CH-rich interacting
match with PLAG1) (Androgen receptor
N-terminal-interacting protein).
Length = 261
Score = 116 bits (290), Expect = 2e-25
Identities = 54/111 (48%), Positives = 69/111 (62%)
Frame = -2
Query: 582 NCPVCMEYLFDSMKAISVLHCGHTIHLECLYEMRAHQQFSCPVCLRSACNMSDIWQKLDQ 403
NCP+C+E + S VL CGH +H C YE + + CP+C+ SA +M+ W++LD
Sbjct: 144 NCPICLEDIHTSRVVAHVLPCGHLLHRTC-YEEMLKEGYRCPLCMHSALDMTRYWRQLDT 202
Query: 402 QVAASPMPAIYQKKMVWILCNDCGVTSNVQFHILAHKCPGCSSYNTRQTRG 250
+VA +PMP+ YQ V ILCNDC S VQFHIL KC C SYNT Q G
Sbjct: 203 EVAQTPMPSEYQNVTVDILCNDCNGRSTVQFHILGMKCKLCDSYNTAQAGG 253
>sptr|Q90ZZ0|Q90ZZ0 Zinc finger protein.
Length = 264
Score = 115 bits (289), Expect = 3e-25
Identities = 52/118 (44%), Positives = 69/118 (58%), Gaps = 3/118 (2%)
Frame = -2
Query: 582 NCPVCMEYLFDSMKAISVLHCGHTIHLECLYEMRAHQQFSCPVCLRSACNMSDIWQKLDQ 403
NCPVCME + VL CGH +H C +M + CP+C+ SA NM + W+++D+
Sbjct: 132 NCPVCMEDIHTFRVGAHVLPCGHLLHGTCFDDMLKTGAYRCPLCMHSAFNMKEYWKQMDE 191
Query: 402 QVAASPMPAIYQKKMVWILCNDCGVTSNVQFHILAHKCPGCSSYNTRQ---TRGDPAA 238
+++ +PMP YQ V I+CNDC S V FH+L KC C SYNT Q G PAA
Sbjct: 192 EISQTPMPTEYQDSTVKIICNDCQARSTVSFHVLGMKCSSCGSYNTAQDGGLMGTPAA 249
>sptr|Q9LYC4|Q9LYC4 Hypothetical 30.9 kDa protein.
Length = 274
Score = 115 bits (287), Expect = 5e-25
Identities = 54/118 (45%), Positives = 70/118 (59%), Gaps = 2/118 (1%)
Frame = -2
Query: 585 HNCPVCMEYLFDSMKAISVLHCGHTIHLECLYEMRAHQQFSCPVCLRSACNMSDIWQKLD 406
++CPVC EYLFDS+KA V+ CGHT+H++C +M Q+ CP+C +S +MS W LD
Sbjct: 168 NSCPVCYEYLFDSVKAAHVMKCGHTMHMDCFEQMINENQYRCPICAKSMVDMSPSWHLLD 227
Query: 405 QQVAASPMPAIYQKKMVWILCNDCGVTSNVQFHILAHKCPGCSSYNTRQ--TRGDPAA 238
+V+ ILCNDC S FHIL HKC C SYNTR+ T DP +
Sbjct: 228 FEVS--------------ILCNDCNKGSKAMFHILGHKCSDCGSYNTRRISTPQDPVS 271
>sptr|Q96BL1|Q96BL1 Hypothetical protein (Fragment).
Length = 117
Score = 108 bits (269), Expect = 6e-23
Identities = 51/111 (45%), Positives = 66/111 (59%)
Frame = -2
Query: 582 NCPVCMEYLFDSMKAISVLHCGHTIHLECLYEMRAHQQFSCPVCLRSACNMSDIWQKLDQ 403
NCP+C+E + S VL CGH +H + + CP+C+ SA +M+ W++LD
Sbjct: 9 NCPICLEDIHTSRVVAHVLPCGHLLH----------RGYRCPLCMHSALDMTRYWRQLDD 58
Query: 402 QVAASPMPAIYQKKMVWILCNDCGVTSNVQFHILAHKCPGCSSYNTRQTRG 250
+VA +PMP+ YQ V ILCNDC S VQFHIL KC C SYNT Q G
Sbjct: 59 EVAQTPMPSEYQNMTVDILCNDCNGRSTVQFHILGMKCKICESYNTAQAGG 109
>sptrnew|EAA15075|EAA15075 ENSANGP00000012255 (Fragment).
Length = 578
Score = 103 bits (257), Expect = 1e-21
Identities = 44/110 (40%), Positives = 65/110 (59%)
Frame = -2
Query: 582 NCPVCMEYLFDSMKAISVLHCGHTIHLECLYEMRAHQQFSCPVCLRSACNMSDIWQKLDQ 403
NCPVC++ + S + CGH +H C E+ + ++CP C S +M+ +W+ LD
Sbjct: 220 NCPVCLDDIHTSRIPCHIPDCGHLLHRTCFEELLSSGHYACPTCQTSMMDMNQLWEYLDA 279
Query: 402 QVAASPMPAIYQKKMVWILCNDCGVTSNVQFHILAHKCPGCSSYNTRQTR 253
+VAA+PMP Y V ILC DC S V+FH++ KC C +YNT +T+
Sbjct: 280 EVAATPMPKEYANYFVDILCKDCHKESTVKFHVVGLKCTHCGAYNTCRTK 329
>sptr|Q8S9V3|Q8S9V3 Putative zinc finger protein.
Length = 1063
Score = 102 bits (253), Expect = 4e-21
Identities = 47/117 (40%), Positives = 67/117 (57%)
Frame = -2
Query: 582 NCPVCMEYLFDSMKAISVLHCGHTIHLECLYEMRAHQQFSCPVCLRSACNMSDIWQKLDQ 403
NCP+C ++LF S A+ L CGH +H C ++ ++CP+C +S +M+ + LD
Sbjct: 945 NCPICCDFLFTSSAAVRALPCGHFMHSAC-FQAYTCSHYTCPICCKSLGDMAVYFGMLDA 1003
Query: 402 QVAASPMPAIYQKKMVWILCNDCGVTSNVQFHILAHKCPGCSSYNTRQTRGDPAACS 232
+AA +P Y+ + ILCNDC +FH L HKC C SYNTR + D A CS
Sbjct: 1004 LLAAEELPEEYRDRCQDILCNDCERKGRSRFHWLYHKCGSCGSYNTRVIKTDTADCS 1060
>sptrnew|EAA34290|EAA34290 Hypothetical protein.
Length = 888
Score = 101 bits (252), Expect = 6e-21
Identities = 44/106 (41%), Positives = 62/106 (58%)
Frame = -2
Query: 582 NCPVCMEYLFDSMKAISVLHCGHTIHLECLYEMRAHQQFSCPVCLRSACNMSDIWQKLDQ 403
+CP+C +YLF+S + ++++ CGHTIH C + + + CPVC +S NM ++ LD
Sbjct: 483 DCPICGDYLFNSPRPVAIMKCGHTIHKHC-FSAHRERSYRCPVCNKSCVNMEIQFRNLDI 541
Query: 402 QVAASPMPAIYQKKMVWILCNDCGVTSNVQFHILAHKCPGCSSYNT 265
+A PMP YQ I CNDC S ++H L KC C SYNT
Sbjct: 542 AIATQPMPDEYQDARAVISCNDCSAKSQTRYHWLGLKCSVCHSYNT 587
>sptr|Q873B9|Q873B9 Hypothetical protein B24N11.050.
Length = 888
Score = 101 bits (252), Expect = 6e-21
Identities = 44/106 (41%), Positives = 62/106 (58%)
Frame = -2
Query: 582 NCPVCMEYLFDSMKAISVLHCGHTIHLECLYEMRAHQQFSCPVCLRSACNMSDIWQKLDQ 403
+CP+C +YLF+S + ++++ CGHTIH C + + + CPVC +S NM ++ LD
Sbjct: 483 DCPICGDYLFNSPRPVAIMKCGHTIHKHC-FSAHRERSYRCPVCNKSCVNMEIQFRNLDI 541
Query: 402 QVAASPMPAIYQKKMVWILCNDCGVTSNVQFHILAHKCPGCSSYNT 265
+A PMP YQ I CNDC S ++H L KC C SYNT
Sbjct: 542 AIATQPMPDEYQDARAVISCNDCSAKSQTRYHWLGLKCSVCHSYNT 587
>sptr|Q9SSG0|Q9SSG0 F25A4.27 protein.
Length = 255
Score = 101 bits (251), Expect = 7e-21
Identities = 46/110 (41%), Positives = 60/110 (54%)
Frame = -2
Query: 591 MHHNCPVCMEYLFDSMKAISVLHCGHTIHLECLYEMRAHQQFSCPVCLRSACNMSDIWQK 412
+ NCP+C EY+F S + L CGH +H C E ++CPVC +S +M ++
Sbjct: 145 LEDNCPICHEYIFTSSSPVKALPCGHLMHSTCFQEYTC-SHYTCPVCSKSLGDMQVYFKM 203
Query: 411 LDQQVAASPMPAIYQKKMVWILCNDCGVTSNVQFHILAHKCPGCSSYNTR 262
LD +A MP Y K ILCNDCG N +H L HKC C SYN+R
Sbjct: 204 LDALLAEEKMPDEYSNKTQVILCNDCGRKGNAPYHWLYHKCTTCGSYNSR 253
>sptr|Q9LMD9|Q9LMD9 F14D16.3.
Length = 1260
Score = 101 bits (251), Expect = 7e-21
Identities = 45/110 (40%), Positives = 61/110 (55%)
Frame = -2
Query: 591 MHHNCPVCMEYLFDSMKAISVLHCGHTIHLECLYEMRAHQQFSCPVCLRSACNMSDIWQK 412
+ NCP+C EY+F S + L CGH +H C E ++CP+C +S +M ++
Sbjct: 1150 LEDNCPICHEYIFTSNSPVKALPCGHVMHSTCFQEYTC-SHYTCPICSKSLGDMQVYFRM 1208
Query: 411 LDQQVAASPMPAIYQKKMVWILCNDCGVTSNVQFHILAHKCPGCSSYNTR 262
LD +A MP Y + ILCNDCG N +H L HKC C+SYNTR
Sbjct: 1209 LDALLAEQKMPDEYLNQTQVILCNDCGRKGNAPYHWLYHKCSSCASYNTR 1258
>sptr|Q8LJR1|Q8LJR1 Putative zinc finger protein.
Length = 400
Score = 100 bits (249), Expect = 1e-20
Identities = 46/117 (39%), Positives = 67/117 (57%)
Frame = -2
Query: 582 NCPVCMEYLFDSMKAISVLHCGHTIHLECLYEMRAHQQFSCPVCLRSACNMSDIWQKLDQ 403
NCP+C ++LF S A+ L CGH++H C ++ ++CP+C +S +M+ + LD
Sbjct: 282 NCPICCDFLFTSSAAVRALPCGHSMHSAC-FQAYTCSHYTCPICCKSLGDMAVYFGMLDA 340
Query: 402 QVAASPMPAIYQKKMVWILCNDCGVTSNVQFHILAHKCPGCSSYNTRQTRGDPAACS 232
+AA +P Y+ + ILCNDC +FH L HKC C SYNTR + A CS
Sbjct: 341 LLAAEELPEEYRDRCQDILCNDCERKGRCRFHWLYHKCGSCGSYNTRVIKTATADCS 397
>sptr|Q9LJQ1|Q9LJQ1 Gb|AAD55299.1.
Length = 1232
Score = 99.4 bits (246), Expect = 3e-20
Identities = 47/119 (39%), Positives = 68/119 (57%), Gaps = 2/119 (1%)
Frame = -2
Query: 582 NCPVCMEYLFDSMKAISVLHCGHTIHLECLYEMRAHQQFSCPVCLRSACNMSDIWQKLDQ 403
NCP+C E+LF S +A+ L CGH +H C ++ ++CP+C +S +M+ + LD
Sbjct: 1112 NCPICCEFLFTSSEAVRALPCGHYMHSAC-FQAYTCSHYTCPICGKSLGDMAVYFGMLDA 1170
Query: 402 QVAASPMPAIYQKKMVWILCNDCGVTSNVQFHILAHKCPGCSSYNTRQTRGD--PAACS 232
+AA +P Y+ + ILCNDC +FH L HKC C SYNTR + + P CS
Sbjct: 1171 LLAAEELPEEYKNRCQDILCNDCERKGTTRFHWLYHKCGSCGSYNTRVIKSETIPPDCS 1229
>sptr|Q8LPQ5|Q8LPQ5 AT3g18290/MIE15_8.
Length = 1254
Score = 99.4 bits (246), Expect = 3e-20
Identities = 47/119 (39%), Positives = 68/119 (57%), Gaps = 2/119 (1%)
Frame = -2
Query: 582 NCPVCMEYLFDSMKAISVLHCGHTIHLECLYEMRAHQQFSCPVCLRSACNMSDIWQKLDQ 403
NCP+C E+LF S +A+ L CGH +H C ++ ++CP+C +S +M+ + LD
Sbjct: 1134 NCPICCEFLFTSSEAVRALPCGHYMHSAC-FQAYTCSHYTCPICGKSLGDMAVYFGMLDA 1192
Query: 402 QVAASPMPAIYQKKMVWILCNDCGVTSNVQFHILAHKCPGCSSYNTRQTRGD--PAACS 232
+AA +P Y+ + ILCNDC +FH L HKC C SYNTR + + P CS
Sbjct: 1193 LLAAEELPEEYKNRCQDILCNDCERKGTTRFHWLYHKCGSCGSYNTRVIKSETIPPDCS 1251
>sptr|O14099|O14099 Zinc finger protein.
Length = 425
Score = 96.3 bits (238), Expect = 2e-19
Identities = 39/106 (36%), Positives = 60/106 (56%)
Frame = -2
Query: 582 NCPVCMEYLFDSMKAISVLHCGHTIHLECLYEMRAHQQFSCPVCLRSACNMSDIWQKLDQ 403
NCP+C EY+F+S + + L C H +H C +E + CP C ++ N++ +++ LD
Sbjct: 270 NCPICGEYMFNSRERVIFLSCSHPLHQRC-HEEYIRTNYRCPTCYKTIINVNSLFRILDM 328
Query: 402 QVAASPMPAIYQKKMVWILCNDCGVTSNVQFHILAHKCPGCSSYNT 265
++ PMP Y + I CNDC + ++H L HKC C SYNT
Sbjct: 329 EIERQPMPYPYNTWISTIRCNDCNSRCDTKYHFLGHKCNSCHSYNT 374
>sptr|Q9VMD4|Q9VMD4 CG16947 protein.
Length = 433
Score = 93.6 bits (231), Expect = 2e-18
Identities = 40/108 (37%), Positives = 58/108 (53%)
Frame = -2
Query: 582 NCPVCMEYLFDSMKAISVLHCGHTIHLECLYEMRAHQQFSCPVCLRSACNMSDIWQKLDQ 403
+CPVC+ + S + CGH +H C ++ A ++CP C S +M+ +W LD
Sbjct: 310 HCPVCLGDIHTSRIPCHIPDCGHLLHKMCFDQLLASGHYTCPTCQTSLIDMTALWVYLDD 369
Query: 402 QVAASPMPAIYQKKMVWILCNDCGVTSNVQFHILAHKCPGCSSYNTRQ 259
Q P+P Y+ + V I CNDC TS +FH + KC C +YNT Q
Sbjct: 370 QAERMPVPLKYENQRVHIFCNDCHKTSKTKFHFIGLKCVHCGAYNTTQ 417
>sptr|Q8SQZ0|Q8SQZ0 LIM domain-containing protein.
Length = 254
Score = 57.0 bits (136), Expect = 2e-07
Identities = 30/101 (29%), Positives = 53/101 (52%)
Frame = -2
Query: 582 NCPVCMEYLFDSMKAISVLHCGHTIHLECLYEMRAHQQFSCPVCLRSACNMSDIWQKLDQ 403
NCP+C E + +SM+ + +L CGH++H C E + ++CP+C + + S I +K++
Sbjct: 135 NCPICAEEMSESMEVLVLLRCGHSLHERCFNEF-IKETYTCPMCSKPIGDTSIINRKVEC 193
Query: 402 QVAASPMPAIYQKKMVWILCNDCGVTSNVQFHILAHKCPGC 280
+ P P+ Q C +C S + + +KCP C
Sbjct: 194 LLGMEP-PSPEQSPKNIAKCTNCNNLS--KCGAIQNKCPFC 231
>sptrnew|EAA38361|EAA38361 GLP_375_16333_15149.
Length = 394
Score = 50.8 bits (120), Expect = 1e-05
Identities = 35/115 (30%), Positives = 54/115 (46%), Gaps = 7/115 (6%)
Frame = -2
Query: 582 NCPVCMEYLFDSMKAISVLHCG-HTIHLECLYEM---RAHQQFSCPVCLRSACNMSD--- 424
NC C++ L D++ VL CG H+ H C YE + CPVC R D
Sbjct: 242 NCVYCLDGLRDNIYPYIVLPCGRHSCHRHC-YEAVFRLGGANYKCPVCRRLVLFGQDREL 300
Query: 423 IWQKLDQQVAASPMPAIYQKKMVWILCNDCGVTSNVQFHILAHKCPGCSSYNTRQ 259
+L++ + +P Y+ V C++CG + H L ++CPGC S+N +
Sbjct: 301 YEARLEELRGSLAVPPEYRGVFVDCACHECGGLFVARRHHL-YRCPGCGSHNAAE 354
>sptrnew|EAA36631|EAA36631 GLP_115_10715_10044.
Length = 223
Score = 50.8 bits (120), Expect = 1e-05
Identities = 35/115 (30%), Positives = 54/115 (46%), Gaps = 7/115 (6%)
Frame = -2
Query: 582 NCPVCMEYLFDSMKAISVLHCG-HTIHLECLYEM---RAHQQFSCPVCLRSACNMSD--- 424
NC C++ L D++ VL CG H+ H C YE + CPVC R D
Sbjct: 67 NCVYCLDGLRDNIYPYIVLPCGRHSCHRHC-YEAVFRLGGANYKCPVCRRLVLFGQDREL 125
Query: 423 IWQKLDQQVAASPMPAIYQKKMVWILCNDCGVTSNVQFHILAHKCPGCSSYNTRQ 259
+L++ + +P Y+ V C++CG + H L ++CPGC S+N +
Sbjct: 126 YEARLEELRGSLAVPPEYRGVFVDCACHECGGLFVARRHHL-YRCPGCGSHNAAE 179
>sptr|Q84P60|Q84P60 Putative zinc finger protein (Fragment).
Length = 407
Score = 49.7 bits (117), Expect = 3e-05
Identities = 19/52 (36%), Positives = 32/52 (61%)
Frame = -2
Query: 582 NCPVCMEYLFDSMKAISVLHCGHTIHLECLYEMRAHQQFSCPVCLRSACNMS 427
NCP+C ++LF S A+ L CGH +H C ++ ++CP+C +S +M+
Sbjct: 246 NCPICCDFLFTSSAAVKGLPCGHFMHSAC-FQAYTCSHYTCPICSKSLGDMT 296
Database: /db/trembl-ebi/tmp/swall
Posted date: Sep 26, 2003 8:29 PM
Number of letters in database: 406,886,216
Number of sequences in database: 1,272,877
Lambda K H
0.318 0.135 0.401
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 511,503,605
Number of Sequences: 1272877
Number of extensions: 11598709
Number of successful extensions: 34117
Number of sequences better than 10.0: 30
Number of HSP's better than 10.0 without gapping: 31516
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 34002
length of database: 406,886,216
effective HSP length: 116
effective length of database: 259,232,484
effective search space used: 20738598720
frameshift window, decay const: 50, 0.1
T: 12
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)

