BLASTX 2.2.6 [Apr-09-2003]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= 3438088.2.1
(615 letters)
Database: /db/uniprot/tmp/swall
1,395,590 sequences; 442,889,342 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
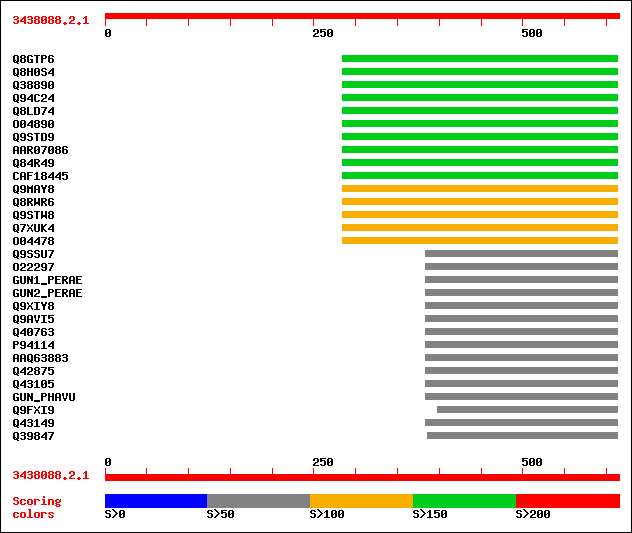
sptr|Q8GTP6|Q8GTP6 Endo-1,4-beta-D-glucanase (EC 3.2.1.4). 155 3e-37
sptr|Q8H0S4|Q8H0S4 At5g49720/K2I5_8. 155 4e-37
sptr|Q38890|Q38890 Cellulase precursor (Cellulase homolog OR16PE... 155 4e-37
sptr|Q94C24|Q94C24 AT5g49720/K2I5_8. 155 4e-37
sptr|Q8LD74|Q8LD74 Cellulase homolog OR16pep. 155 4e-37
sptr|O04890|O04890 Endo-1,4-beta-glucanase (EC 3.2.1.4). 154 7e-37
sptr|Q9STD9|Q9STD9 Cellulase (EC 3.2.1.4). 154 9e-37
sptrnew|AAR07086|AAR07086 Putative endo-1,4-beta-glucanase. 153 1e-36
sptr|Q84R49|Q84R49 Putative endo-1,4-beta-glucanase. 153 1e-36
sptrnew|CAF18445|CAF18445 Endo-1,4-beta-D-glucanase KORRIGAN (EC... 151 6e-36
sptr|Q9MAY8|Q9MAY8 Endo-1,4-beta-glucanase Cel1. 150 1e-35
sptr|Q8RWR6|Q8RWR6 Endo-1,4-beta-glucanase. 149 2e-35
sptr|Q9STW8|Q9STW8 Endo-1, 4-beta-glucanase like protein. 134 9e-31
sptr|Q7XUK4|Q7XUK4 OSJNBa0067K08.14 protein. 125 4e-28
sptr|O04478|O04478 F5I14.14. 117 9e-26
sptr|Q9SSU7|Q9SSU7 Endo-1,4-beta-glucanase. 96 4e-19
sptr|O22297|O22297 Acidic cellulase. 94 1e-18
sw|P05522|GUN1_PERAE Endoglucanase 1 precursor (EC 3.2.1.4) (End... 94 2e-18
sw|P23666|GUN2_PERAE Endoglucanase 2 (EC 3.2.1.4) (Endo-1,4-beta... 92 5e-18
sptr|Q9XIY8|Q9XIY8 Endo-1,4-beta glucanase precursor. 92 5e-18
sptr|Q9AVI5|Q9AVI5 Endo-1,4-beta glucanase precursor. 92 5e-18
sptr|Q40763|Q40763 Cellulase precursor. 92 7e-18
sptr|P94114|P94114 Endo-beta-1,4-glucanase (EC 3.2.1.4) (Cellula... 91 9e-18
sptrnew|AAQ63883|AAQ63883 Cellulase. 91 1e-17
sptr|Q42875|Q42875 Endo-1,4-beta-glucanase precursor (EC 3.2.1.4). 91 2e-17
sptr|Q43105|Q43105 Cellulase (EC 3.2.1.4). 91 2e-17
sw|P22503|GUN_PHAVU Endoglucanase precursor (EC 3.2.1.4) (Endo-1... 91 2e-17
sptr|Q9FXI9|Q9FXI9 F6F9.1 protein (At1g19940/F6F9_1). 90 3e-17
sptr|Q43149|Q43149 Cellulase (EC 3.2.1.4) (Fragment). 89 3e-17
sptr|Q39847|Q39847 CMCase, cellulase, endo-1,4-beta-D-glucanase ... 89 3e-17
>sptr|Q8GTP6|Q8GTP6 Endo-1,4-beta-D-glucanase (EC 3.2.1.4).
Length = 621
Score = 155 bits (393), Expect = 3e-37
Identities = 70/110 (63%), Positives = 76/110 (69%)
Frame = -3
Query: 613 NPLKMSYVVGFGKRYPRHVHHRGASIPKNGVHYGCKGGWKWRDAKKPNPNTIAGAMVAGP 434
NP KMSYVVGFG YP+HVHHRGASIPKN + Y CKGGWKWRD K NPNTI GAMVAGP
Sbjct: 496 NPRKMSYVVGFGNHYPKHVHHRGASIPKNKIKYSCKGGWKWRDTPKANPNTIDGAMVAGP 555
Query: 433 DRHDGFKDARRNYNYTEXXXXXXXXXXXXXXXXXXXGHGGVDKNTMFSAV 284
D+HDGF+D R NYNYTE G+DKNT+FSAV
Sbjct: 556 DKHDGFRDVRSNYNYTEPTLAGNAGLVAALVALSGEKSVGIDKNTIFSAV 605
>sptr|Q8H0S4|Q8H0S4 At5g49720/K2I5_8.
Length = 621
Score = 155 bits (392), Expect = 4e-37
Identities = 72/112 (64%), Positives = 79/112 (70%), Gaps = 2/112 (1%)
Frame = -3
Query: 613 NPLKMSYVVGFGKRYPRHVHHRGASIPKNGVHYGCKGGWKWRDAKKPNPNTIAGAMVAGP 434
NP KMSYVVGFG +YPRHVHHRGASIPKN V Y CKGGWKWRD+KKPNPNTI GAMVAGP
Sbjct: 494 NPRKMSYVVGFGTKYPRHVHHRGASIPKNKVKYNCKGGWKWRDSKKPNPNTIEGAMVAGP 553
Query: 433 DRHDGFKDARRNYNYTEXXXXXXXXXXXXXXXXXXXGH--GGVDKNTMFSAV 284
D+ DG++D R NYNYTE G +DKNT+FSAV
Sbjct: 554 DKRDGYRDVRMNYNYTEPTLAGNAGLVAALVALSGEEEATGKIDKNTIFSAV 605
>sptr|Q38890|Q38890 Cellulase precursor (Cellulase homolog OR16PEP
precursor).
Length = 621
Score = 155 bits (392), Expect = 4e-37
Identities = 72/112 (64%), Positives = 79/112 (70%), Gaps = 2/112 (1%)
Frame = -3
Query: 613 NPLKMSYVVGFGKRYPRHVHHRGASIPKNGVHYGCKGGWKWRDAKKPNPNTIAGAMVAGP 434
NP KMSYVVGFG +YPRHVHHRGASIPKN V Y CKGGWKWRD+KKPNPNTI GAMVAGP
Sbjct: 494 NPRKMSYVVGFGTKYPRHVHHRGASIPKNKVKYNCKGGWKWRDSKKPNPNTIEGAMVAGP 553
Query: 433 DRHDGFKDARRNYNYTEXXXXXXXXXXXXXXXXXXXGH--GGVDKNTMFSAV 284
D+ DG++D R NYNYTE G +DKNT+FSAV
Sbjct: 554 DKRDGYRDVRMNYNYTEPTLAGNAGLVAALVALSGEEEATGKIDKNTIFSAV 605
>sptr|Q94C24|Q94C24 AT5g49720/K2I5_8.
Length = 621
Score = 155 bits (392), Expect = 4e-37
Identities = 72/112 (64%), Positives = 79/112 (70%), Gaps = 2/112 (1%)
Frame = -3
Query: 613 NPLKMSYVVGFGKRYPRHVHHRGASIPKNGVHYGCKGGWKWRDAKKPNPNTIAGAMVAGP 434
NP KMSYVVGFG +YPRHVHHRGASIPKN V Y CKGGWKWRD+KKPNPNTI GAMVAGP
Sbjct: 494 NPRKMSYVVGFGTKYPRHVHHRGASIPKNKVKYNCKGGWKWRDSKKPNPNTIEGAMVAGP 553
Query: 433 DRHDGFKDARRNYNYTEXXXXXXXXXXXXXXXXXXXGH--GGVDKNTMFSAV 284
D+ DG++D R NYNYTE G +DKNT+FSAV
Sbjct: 554 DKRDGYRDVRMNYNYTEPTLAGNAGLVAALVALSGEEEATGKIDKNTIFSAV 605
>sptr|Q8LD74|Q8LD74 Cellulase homolog OR16pep.
Length = 621
Score = 155 bits (392), Expect = 4e-37
Identities = 72/112 (64%), Positives = 79/112 (70%), Gaps = 2/112 (1%)
Frame = -3
Query: 613 NPLKMSYVVGFGKRYPRHVHHRGASIPKNGVHYGCKGGWKWRDAKKPNPNTIAGAMVAGP 434
NP KMSYVVGFG +YPRHVHHRGASIPKN V Y CKGGWKWRD+KKPNPNTI GAMVAGP
Sbjct: 494 NPRKMSYVVGFGTKYPRHVHHRGASIPKNKVKYNCKGGWKWRDSKKPNPNTIEGAMVAGP 553
Query: 433 DRHDGFKDARRNYNYTEXXXXXXXXXXXXXXXXXXXGH--GGVDKNTMFSAV 284
D+ DG++D R NYNYTE G +DKNT+FSAV
Sbjct: 554 DKRDGYRDVRMNYNYTEPTLAGNAGLVAALVALSGEEEATGKIDKNTIFSAV 605
>sptr|O04890|O04890 Endo-1,4-beta-glucanase (EC 3.2.1.4).
Length = 617
Score = 154 bits (390), Expect = 7e-37
Identities = 70/110 (63%), Positives = 77/110 (70%)
Frame = -3
Query: 613 NPLKMSYVVGFGKRYPRHVHHRGASIPKNGVHYGCKGGWKWRDAKKPNPNTIAGAMVAGP 434
NP KMSYVVGFG YP+HVHHRGASIPKN V Y CKGGWK+RD+ K NPNTI GAMVAGP
Sbjct: 492 NPRKMSYVVGFGNHYPKHVHHRGASIPKNKVKYNCKGGWKYRDSSKANPNTIVGAMVAGP 551
Query: 433 DRHDGFKDARRNYNYTEXXXXXXXXXXXXXXXXXXXGHGGVDKNTMFSAV 284
D+HDGF+D R NYNYTE G+DKNT+FSAV
Sbjct: 552 DKHDGFRDVRSNYNYTEPTLAGNAGLVAALVALSGDRDVGIDKNTLFSAV 601
>sptr|Q9STD9|Q9STD9 Cellulase (EC 3.2.1.4).
Length = 621
Score = 154 bits (389), Expect = 9e-37
Identities = 71/112 (63%), Positives = 79/112 (70%), Gaps = 2/112 (1%)
Frame = -3
Query: 613 NPLKMSYVVGFGKRYPRHVHHRGASIPKNGVHYGCKGGWKWRDAKKPNPNTIAGAMVAGP 434
NP KMSY+VGFG +YP+HVHHRGASIPKN V Y CKGGWKWRD+KKPNPNTI GAMVAGP
Sbjct: 494 NPRKMSYLVGFGTKYPKHVHHRGASIPKNKVKYNCKGGWKWRDSKKPNPNTIEGAMVAGP 553
Query: 433 DRHDGFKDARRNYNYTE--XXXXXXXXXXXXXXXXXXXGHGGVDKNTMFSAV 284
D+ DGF+D R NYNYTE G +DKNT+FSAV
Sbjct: 554 DKRDGFRDVRTNYNYTEPTLAGNAGLVAALVALSGEEEASGTIDKNTIFSAV 605
>sptrnew|AAR07086|AAR07086 Putative endo-1,4-beta-glucanase.
Length = 620
Score = 153 bits (387), Expect = 1e-36
Identities = 70/111 (63%), Positives = 78/111 (70%), Gaps = 1/111 (0%)
Frame = -3
Query: 613 NPLKMSYVVGFGKRYPRHVHHRGASIPKNGVHYGCKGGWKWRDAKKPNPNTIAGAMVAGP 434
NPLKMSYVVGFG +YP+ HHRGASIP NGV YGCKGG+KWR+ KKPNPN + GA+VAGP
Sbjct: 494 NPLKMSYVVGFGNKYPKRAHHRGASIPHNGVKYGCKGGFKWRETKKPNPNILIGALVAGP 553
Query: 433 DRHDGFKDARRNYNYTE-XXXXXXXXXXXXXXXXXXXGHGGVDKNTMFSAV 284
DRHDGFKD R NYNYTE G+DKNT+FSAV
Sbjct: 554 DRHDGFKDVRTNYNYTEPTLAANAGLVAALISLTNIHVKSGIDKNTIFSAV 604
>sptr|Q84R49|Q84R49 Putative endo-1,4-beta-glucanase.
Length = 620
Score = 153 bits (387), Expect = 1e-36
Identities = 70/111 (63%), Positives = 78/111 (70%), Gaps = 1/111 (0%)
Frame = -3
Query: 613 NPLKMSYVVGFGKRYPRHVHHRGASIPKNGVHYGCKGGWKWRDAKKPNPNTIAGAMVAGP 434
NPLKMSYVVGFG +YP+ HHRGASIP NGV YGCKGG+KWR+ KKPNPN + GA+VAGP
Sbjct: 494 NPLKMSYVVGFGNKYPKRAHHRGASIPHNGVKYGCKGGFKWRETKKPNPNILIGALVAGP 553
Query: 433 DRHDGFKDARRNYNYTE-XXXXXXXXXXXXXXXXXXXGHGGVDKNTMFSAV 284
DRHDGFKD R NYNYTE G+DKNT+FSAV
Sbjct: 554 DRHDGFKDVRTNYNYTEPTLAANAGLVAALISLTNIHVKSGIDKNTIFSAV 604
>sptrnew|CAF18445|CAF18445 Endo-1,4-beta-D-glucanase KORRIGAN (EC
3.2.1.4) (Fragment).
Length = 229
Score = 151 bits (382), Expect = 6e-36
Identities = 68/110 (61%), Positives = 73/110 (66%)
Frame = -3
Query: 613 NPLKMSYVVGFGKRYPRHVHHRGASIPKNGVHYGCKGGWKWRDAKKPNPNTIAGAMVAGP 434
NP KMSY+VGFG YP+HVHHRGASIPK V Y CKGGWKWRD KPNPN + GAMVAGP
Sbjct: 111 NPRKMSYIVGFGNHYPKHVHHRGASIPKTKVKYNCKGGWKWRDTSKPNPNILVGAMVAGP 170
Query: 433 DRHDGFKDARRNYNYTEXXXXXXXXXXXXXXXXXXXGHGGVDKNTMFSAV 284
DRHDGF D R NYNYTE +DKNT+FSAV
Sbjct: 171 DRHDGFHDVRSNYNYTEPTLAGNAGLVAALVALSGDKSIPIDKNTLFSAV 220
>sptr|Q9MAY8|Q9MAY8 Endo-1,4-beta-glucanase Cel1.
Length = 621
Score = 150 bits (380), Expect = 1e-35
Identities = 70/112 (62%), Positives = 79/112 (70%), Gaps = 2/112 (1%)
Frame = -3
Query: 613 NPLKMSYVVGFGKRYPRHVHHRGASIPKNGVHYGCKGGWKWRDAKKPNPNTIAGAMVAGP 434
NP KMSYVVGFGK+YP+ VHHRGASIP NGV YGCKGG+KWR++KK NPN + GAMVAGP
Sbjct: 494 NPQKMSYVVGFGKKYPKRVHHRGASIPHNGVKYGCKGGFKWRESKKANPNILVGAMVAGP 553
Query: 433 DRHDGFKDARRNYNYTEXXXXXXXXXXXXXXXXXXXGHG--GVDKNTMFSAV 284
D+HDGFKD R NYNYTE G +DKNT+FSAV
Sbjct: 554 DKHDGFKDIRTNYNYTEPTLAANAGLVAALISLADIDTGRYSIDKNTIFSAV 605
>sptr|Q8RWR6|Q8RWR6 Endo-1,4-beta-glucanase.
Length = 620
Score = 149 bits (377), Expect = 2e-35
Identities = 70/112 (62%), Positives = 78/112 (69%), Gaps = 2/112 (1%)
Frame = -3
Query: 613 NPLKMSYVVGFGKRYPRHVHHRGASIPKNGVHYGCKGGWKWRDAKKPNPNTIAGAMVAGP 434
NP KMSYVVGFGK+YP+ VHHRGASIP NGV YGCKGG+KWR+ KK NPN + GAMVAGP
Sbjct: 494 NPQKMSYVVGFGKKYPKRVHHRGASIPHNGVKYGCKGGFKWREFKKANPNILVGAMVAGP 553
Query: 433 DRHDGFKDARRNYNYTEXXXXXXXXXXXXXXXXXXXGHG--GVDKNTMFSAV 284
D+HDGFKD R NYNYTE G +DKNT+FSAV
Sbjct: 554 DKHDGFKDIRTNYNYTEPTLAANAGLVAALISLADIDTGRYSIDKNTIFSAV 605
>sptr|Q9STW8|Q9STW8 Endo-1, 4-beta-glucanase like protein.
Length = 620
Score = 134 bits (337), Expect = 9e-31
Identities = 66/111 (59%), Positives = 75/111 (67%), Gaps = 1/111 (0%)
Frame = -3
Query: 613 NPLKMSYVVGFGKRYPRHVHHRGASIPKNGVHYGCKGGWKWRDAKKPNPNTIAGAMVAGP 434
NP KMSYVVG+G+RYP+ VHHRGASIPKN + C GG+KW+ +KK NPN I GAMVAGP
Sbjct: 495 NPRKMSYVVGYGQRYPKQVHHRGASIPKN-MKETCTGGFKWKKSKKNNPNAINGAMVAGP 553
Query: 433 DRHDGFKDARRNYNYTE-XXXXXXXXXXXXXXXXXXXGHGGVDKNTMFSAV 284
D+HDGF D R NYNYTE GG+DKNTMFSAV
Sbjct: 554 DKHDGFHDIRTNYNYTEPTLAGNAGLVAALVALSGEKAVGGIDKNTMFSAV 604
>sptr|Q7XUK4|Q7XUK4 OSJNBa0067K08.14 protein.
Length = 623
Score = 125 bits (314), Expect = 4e-28
Identities = 61/114 (53%), Positives = 70/114 (61%), Gaps = 4/114 (3%)
Frame = -3
Query: 613 NPLKMSYVVGFGKRYPRHVHHRGASIPKNGVHYGCKGGWKWRDAKKPNPNTIAGAMVAGP 434
NP KMSYVVG+GK+YPR +HHRGAS P NG+ Y C GG+KWRD K +PN + GAMV GP
Sbjct: 494 NPKKMSYVVGYGKKYPRRLHHRGASTPHNGIKYSCTGGYKWRDTKGADPNVLVGAMVGGP 553
Query: 433 DRHDGFKDARRNYNYTEXXXXXXXXXXXXXXXXXXXGHG----GVDKNTMFSAV 284
D++D FKDAR Y E G G VDKNTMFSAV
Sbjct: 554 DKNDQFKDARLTYAQNEPTLVGNAGLVAALVALTNSGRGAGVTAVDKNTMFSAV 607
>sptr|O04478|O04478 F5I14.14.
Length = 623
Score = 117 bits (294), Expect = 9e-26
Identities = 56/110 (50%), Positives = 68/110 (61%)
Frame = -3
Query: 613 NPLKMSYVVGFGKRYPRHVHHRGASIPKNGVHYGCKGGWKWRDAKKPNPNTIAGAMVAGP 434
NPLKMSYVVGFGK++PR VHHRGA+IP + C+ G K+RD K PNPN I GAMV GP
Sbjct: 498 NPLKMSYVVGFGKKFPRRVHHRGATIPNDKKRRSCREGLKYRDTKNPNPNNITGAMVGGP 557
Query: 433 DRHDGFKDARRNYNYTEXXXXXXXXXXXXXXXXXXXGHGGVDKNTMFSAV 284
++ D F D R NYN +E G +DKNTMF++V
Sbjct: 558 NKFDEFHDLRNNYNASEPTLSGNAGLVAALVSLTSSGGQQIDKNTMFNSV 607
>sptr|Q9SSU7|Q9SSU7 Endo-1,4-beta-glucanase.
Length = 506
Score = 95.9 bits (237), Expect = 4e-19
Identities = 42/80 (52%), Positives = 56/80 (70%), Gaps = 3/80 (3%)
Frame = -3
Query: 613 NPLKMSYVVGFGKRYPRHVHHRGASIPKNGVHYG---CKGGWKWRDAKKPNPNTIAGAMV 443
NPLKMSY+VG+G RYP+ +HHRG+S+P VH G C G+ ++K PNPN + GA+V
Sbjct: 404 NPLKMSYMVGYGPRYPQRIHHRGSSLPSMAVHPGKIQCSAGFGVMNSKSPNPNILMGAVV 463
Query: 442 AGPDRHDGFKDARRNYNYTE 383
GPD+HD F D R +Y +E
Sbjct: 464 GGPDQHDRFPDQRSDYEQSE 483
>sptr|O22297|O22297 Acidic cellulase.
Length = 505
Score = 94.0 bits (232), Expect = 1e-18
Identities = 40/80 (50%), Positives = 55/80 (68%), Gaps = 3/80 (3%)
Frame = -3
Query: 613 NPLKMSYVVGFGKRYPRHVHHRGASIPK---NGVHYGCKGGWKWRDAKKPNPNTIAGAMV 443
NP KMSY+VGFG+RYP+HVHHRG+S+P + H C G+++ ++ PNPN + GA++
Sbjct: 403 NPAKMSYMVGFGERYPQHVHHRGSSLPSIHAHPDHIACNDGFQYLYSRSPNPNVLTGAIL 462
Query: 442 AGPDRHDGFKDARRNYNYTE 383
GPD D F D R NY +E
Sbjct: 463 GGPDNRDNFADDRNNYQQSE 482
>sw|P05522|GUN1_PERAE Endoglucanase 1 precursor (EC 3.2.1.4)
(Endo-1,4-beta-glucanase) (Abscission cellulase 1).
Length = 494
Score = 93.6 bits (231), Expect = 2e-18
Identities = 41/80 (51%), Positives = 54/80 (67%), Gaps = 3/80 (3%)
Frame = -3
Query: 613 NPLKMSYVVGFGKRYPRHVHHRGASIPKNGVH---YGCKGGWKWRDAKKPNPNTIAGAMV 443
NP KMSY+VGFG+RYP+HVHHRG+S+P VH C G+++ + PNPN + GA++
Sbjct: 392 NPAKMSYMVGFGERYPQHVHHRGSSLPSVQVHPNSIPCNAGFQYLYSSPPNPNILVGAIL 451
Query: 442 AGPDRHDGFKDARRNYNYTE 383
GPD D F D R NY +E
Sbjct: 452 GGPDNRDSFSDDRNNYQQSE 471
>sw|P23666|GUN2_PERAE Endoglucanase 2 (EC 3.2.1.4)
(Endo-1,4-beta-glucanase) (Abscission cellulase 2)
(Fragment).
Length = 130
Score = 92.0 bits (227), Expect = 5e-18
Identities = 40/80 (50%), Positives = 53/80 (66%), Gaps = 3/80 (3%)
Frame = -3
Query: 613 NPLKMSYVVGFGKRYPRHVHHRGASIPKNGVH---YGCKGGWKWRDAKKPNPNTIAGAMV 443
NP KMSY+VGFG+RYP+HVHHRG+S+P H C G+++ + PNPN + GA++
Sbjct: 28 NPAKMSYMVGFGERYPQHVHHRGSSLPSVHAHPNPIPCNAGFQYLYSSSPNPNILVGAIL 87
Query: 442 AGPDRHDGFKDARRNYNYTE 383
GPD D F D R NY +E
Sbjct: 88 GGPDSRDSFSDDRNNYQQSE 107
>sptr|Q9XIY8|Q9XIY8 Endo-1,4-beta glucanase precursor.
Length = 494
Score = 92.0 bits (227), Expect = 5e-18
Identities = 40/80 (50%), Positives = 52/80 (65%), Gaps = 3/80 (3%)
Frame = -3
Query: 613 NPLKMSYVVGFGKRYPRHVHHRGASIPKNGVH---YGCKGGWKWRDAKKPNPNTIAGAMV 443
NP KMSY+VGFG +YP+HVHHRG+S+P H C G+++ + PNPN + GA+V
Sbjct: 395 NPAKMSYMVGFGNKYPQHVHHRGSSVPSIHAHPNRISCNDGFQYLYSSSPNPNVLVGAIV 454
Query: 442 AGPDRHDGFKDARRNYNYTE 383
GPD D F D R NY +E
Sbjct: 455 GGPDNRDHFADDRNNYQQSE 474
>sptr|Q9AVI5|Q9AVI5 Endo-1,4-beta glucanase precursor.
Length = 494
Score = 92.0 bits (227), Expect = 5e-18
Identities = 39/80 (48%), Positives = 52/80 (65%), Gaps = 3/80 (3%)
Frame = -3
Query: 613 NPLKMSYVVGFGKRYPRHVHHRGASIPKNGVH---YGCKGGWKWRDAKKPNPNTIAGAMV 443
NP +MSY+VGFG RYP+HVHHRG+S+P H C G+++ + PNPN + GA++
Sbjct: 395 NPARMSYMVGFGNRYPQHVHHRGSSVPSIHAHPNRISCNDGFQFLYSSSPNPNVLVGAII 454
Query: 442 AGPDRHDGFKDARRNYNYTE 383
GPD D F D R NY +E
Sbjct: 455 GGPDNRDNFADDRNNYQQSE 474
>sptr|Q40763|Q40763 Cellulase precursor.
Length = 494
Score = 91.7 bits (226), Expect = 7e-18
Identities = 38/80 (47%), Positives = 53/80 (66%), Gaps = 3/80 (3%)
Frame = -3
Query: 613 NPLKMSYVVGFGKRYPRHVHHRGASIPK---NGVHYGCKGGWKWRDAKKPNPNTIAGAMV 443
NP +MSY+VGFG RYP+HVHHRG+S+P + C G+++ + PNPN + GA++
Sbjct: 395 NPARMSYMVGFGNRYPQHVHHRGSSVPSIHDTRIGISCNDGFQFLYSSSPNPNVLVGAII 454
Query: 442 AGPDRHDGFKDARRNYNYTE 383
GPD D F D+R NY +E
Sbjct: 455 GGPDNRDNFADSRNNYQQSE 474
>sptr|P94114|P94114 Endo-beta-1,4-glucanase (EC 3.2.1.4) (Cellulase)
(Endoglucanase) (Endo-1,4-beta-glucanase) (Carboxymethyl
cellulase).
Length = 497
Score = 91.3 bits (225), Expect = 9e-18
Identities = 39/80 (48%), Positives = 53/80 (66%), Gaps = 3/80 (3%)
Frame = -3
Query: 613 NPLKMSYVVGFGKRYPRHVHHRGASIPKNGVH---YGCKGGWKWRDAKKPNPNTIAGAMV 443
NP K+SY+VGFGK+YP H+HHRG+S+P H C G+++ ++ PNPN + GA+V
Sbjct: 395 NPAKISYMVGFGKKYPLHIHHRGSSLPSVHEHPERISCNNGFQYLNSGSPNPNVLVGAIV 454
Query: 442 AGPDRHDGFKDARRNYNYTE 383
GPD D F D R NY +E
Sbjct: 455 GGPDSKDSFSDDRNNYQQSE 474
>sptrnew|AAQ63883|AAQ63883 Cellulase.
Length = 601
Score = 90.9 bits (224), Expect = 1e-17
Identities = 40/77 (51%), Positives = 51/77 (66%)
Frame = -3
Query: 613 NPLKMSYVVGFGKRYPRHVHHRGASIPKNGVHYGCKGGWKWRDAKKPNPNTIAGAMVAGP 434
NP+KMSY+VG+G ++P VHHR ASIP + Y C G W ++K PNP + GAMV GP
Sbjct: 491 NPMKMSYLVGYGDKFPVQVHHRSASIPWDKRLYNCDDGKTWLNSKNPNPQVLLGAMVGGP 550
Query: 433 DRHDGFKDARRNYNYTE 383
D +D F D R N +TE
Sbjct: 551 DTNDHFTDQRSNKRFTE 567
>sptr|Q42875|Q42875 Endo-1,4-beta-glucanase precursor (EC 3.2.1.4).
Length = 510
Score = 90.5 bits (223), Expect = 2e-17
Identities = 39/80 (48%), Positives = 54/80 (67%), Gaps = 3/80 (3%)
Frame = -3
Query: 613 NPLKMSYVVGFGKRYPRHVHHRGASIPKNGVH---YGCKGGWKWRDAKKPNPNTIAGAMV 443
NPLKMSY+VG+G RYP+ +HHRG+S+P H C+ G+ +++ PNPN + GA+V
Sbjct: 408 NPLKMSYMVGYGARYPQRIHHRGSSLPSVANHPAKIQCRDGFSVMNSQSPNPNVLVGAVV 467
Query: 442 AGPDRHDGFKDARRNYNYTE 383
GPD HD F D R +Y +E
Sbjct: 468 GGPDEHDRFPDERSDYEQSE 487
>sptr|Q43105|Q43105 Cellulase (EC 3.2.1.4).
Length = 496
Score = 90.5 bits (223), Expect = 2e-17
Identities = 42/81 (51%), Positives = 56/81 (69%), Gaps = 4/81 (4%)
Frame = -3
Query: 613 NPLKMSYVVGFGKRYPRHVHHRGASIPKNGVH---YGCKGGWK-WRDAKKPNPNTIAGAM 446
NP+KMSY+VGFG +YP+ +HHRG+SIP VH GC G + ++ PNPNT GA+
Sbjct: 396 NPMKMSYMVGFGSKYPKQLHHRGSSIPSIKVHPAKVGCNAGLSDYYNSANPNPNTHVGAI 455
Query: 445 VAGPDRHDGFKDARRNYNYTE 383
V GPD +D F DAR +Y++ E
Sbjct: 456 VGGPDSNDRFNDARSDYSHAE 476
>sw|P22503|GUN_PHAVU Endoglucanase precursor (EC 3.2.1.4)
(Endo-1,4-beta-glucanase) (Abscission cellulase).
Length = 496
Score = 90.5 bits (223), Expect = 2e-17
Identities = 42/81 (51%), Positives = 56/81 (69%), Gaps = 4/81 (4%)
Frame = -3
Query: 613 NPLKMSYVVGFGKRYPRHVHHRGASIPKNGVH---YGCKGGWK-WRDAKKPNPNTIAGAM 446
NP+KMSY+VGFG +YP+ +HHRG+SIP VH GC G + ++ PNPNT GA+
Sbjct: 396 NPMKMSYMVGFGSKYPKQLHHRGSSIPSIKVHPAKVGCNAGLSDYYNSANPNPNTHVGAI 455
Query: 445 VAGPDRHDGFKDARRNYNYTE 383
V GPD +D F DAR +Y++ E
Sbjct: 456 VGGPDSNDRFNDARSDYSHAE 476
>sptr|Q9FXI9|Q9FXI9 F6F9.1 protein (At1g19940/F6F9_1).
Length = 515
Score = 89.7 bits (221), Expect = 3e-17
Identities = 41/72 (56%), Positives = 52/72 (72%)
Frame = -3
Query: 613 NPLKMSYVVGFGKRYPRHVHHRGASIPKNGVHYGCKGGWKWRDAKKPNPNTIAGAMVAGP 434
NP KMSY+VG+G++YP VHHRGASIP + GCK G+KW ++ +PNPN GA+V GP
Sbjct: 414 NPEKMSYLVGYGEKYPEFVHHRGASIPADATT-GCKDGFKWLNSDEPNPNVAYGALVGGP 472
Query: 433 DRHDGFKDARRN 398
+D F DAR N
Sbjct: 473 FLNDTFIDARNN 484
>sptr|Q43149|Q43149 Cellulase (EC 3.2.1.4) (Fragment).
Length = 508
Score = 89.4 bits (220), Expect = 3e-17
Identities = 41/81 (50%), Positives = 54/81 (66%), Gaps = 4/81 (4%)
Frame = -3
Query: 613 NPLKMSYVVGFGKRYPRHVHHRGASIPKNGVH---YGCKGGWK-WRDAKKPNPNTIAGAM 446
NP+KMSY+VGFG +YP +HHRGASIP +H GC G+ W + K NPNT GA+
Sbjct: 389 NPMKMSYMVGFGSKYPLQLHHRGASIPSMQIHPARVGCNEGYSAWYSSSKSNPNTHVGAI 448
Query: 445 VAGPDRHDGFKDARRNYNYTE 383
V GP+ +D F D R +Y++ E
Sbjct: 449 VGGPNSNDQFNDVRSDYSHLE 469
>sptr|Q39847|Q39847 CMCase, cellulase, endo-1,4-beta-D-glucanase
(Fragment).
Length = 299
Score = 89.4 bits (220), Expect = 3e-17
Identities = 41/80 (51%), Positives = 56/80 (70%), Gaps = 4/80 (5%)
Frame = -3
Query: 613 NPLKMSYVVGFGKRYPRHVHHRGASIPKNGVH---YGCKGGWK-WRDAKKPNPNTIAGAM 446
NP+KMSY+VGFG +YP+ +HHRG+SIP VH GC G + ++ PNPNT GA+
Sbjct: 183 NPMKMSYMVGFGSKYPKQLHHRGSSIPSINVHPTKVGCNDGLSVYYNSANPNPNTHVGAI 242
Query: 445 VAGPDRHDGFKDARRNYNYT 386
V GPD +D F DAR +Y+++
Sbjct: 243 VGGPDSNDRFSDARSDYSHS 262
Database: /db/uniprot/tmp/swall
Posted date: Mar 5, 2004 7:45 PM
Number of letters in database: 442,889,342
Number of sequences in database: 1,395,590
Lambda K H
0.318 0.135 0.401
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 529,145,430
Number of Sequences: 1395590
Number of extensions: 11662498
Number of successful extensions: 35837
Number of sequences better than 10.0: 30
Number of HSP's better than 10.0 without gapping: 33974
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 35654
length of database: 442,889,342
effective HSP length: 117
effective length of database: 279,605,312
effective search space used: 24325662144
frameshift window, decay const: 50, 0.1
T: 12
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)

