BLASTX 2.2.6 [Apr-09-2003]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= 3336950.2.1
(645 letters)
Database: /db/uniprot/tmp/swall
1,395,590 sequences; 442,889,342 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
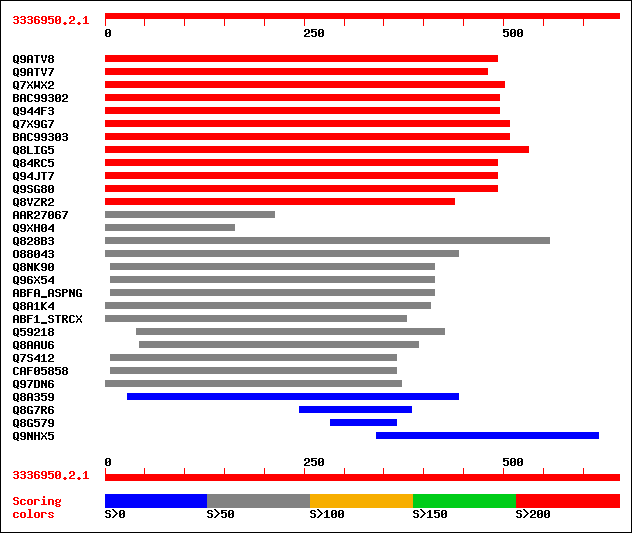
sptr|Q9ATV8|Q9ATV8 Arabinoxylan arabinofuranohydrolase isoenzyme... 291 7e-78
sptr|Q9ATV7|Q9ATV7 Arabinoxylan arabinofuranohydrolase isoenzyme... 290 1e-77
sptr|Q7XWX2|Q7XWX2 OSJNBb0058J09.4 protein. 263 1e-69
sptrnew|BAC99302|BAC99302 Alpha-L-arabinofuranosidase. 233 1e-60
sptr|Q944F3|Q944F3 Arabinosidase ARA-1. 233 1e-60
sptr|Q7X9G7|Q7X9G7 Alpha-L-arabinofuranosidase. 233 2e-60
sptrnew|BAC99303|BAC99303 Alpha-L-arabinofuranosidase. 231 5e-60
sptr|Q8LIG5|Q8LIG5 Putative arabinoxylan narabinofuranohydrolase... 225 3e-58
sptr|Q84RC5|Q84RC5 Alpha-L-arabinofuranosidase. 216 2e-55
sptr|Q94JT7|Q94JT7 AT3g10740/T7M13_18. 216 2e-55
sptr|Q9SG80|Q9SG80 Putative alpha-L-arabinofuranosidase. 216 2e-55
sptr|Q8VZR2|Q8VZR2 Putative arabinosidase (Alpha-L-arabinofurano... 213 1e-54
sptrnew|AAR27067|AAR27067 A-arabinofuranosidase 1 (Fragment). 100 3e-20
sptr|Q9XH04|Q9XH04 T1N24.13 protein. 84 2e-15
sptr|Q828B3|Q828B3 Putative secreted arabinosidase. 70 3e-11
sptr|O88043|O88043 Putative secreted arabinosidase. 66 3e-10
sptr|Q8NK90|Q8NK90 Alpha-L-arabinofuranosidase A (EC 3.2.1.55). 66 3e-10
sptr|Q96X54|Q96X54 Alpha-L-arabinofuranosidase A (EC 3.2.1.55). 66 3e-10
sw|P42254|ABFA_ASPNG Alpha-L-arabinofuranosidase A precursor (EC... 65 8e-10
sptr|Q8A1K4|Q8A1K4 Alpha-L-arabinofuranosidase A precursor. 64 1e-09
sw|P82593|ABF1_STRCX Alpha-L-arabinofuranosidase I precursor (EC... 63 3e-09
sptr|Q59218|Q59218 Arabinosidase (EC 3.2.1.55). 59 5e-08
sptr|Q8AAU6|Q8AAU6 Alpha-L-arabinofuranosidase A precursor. 59 7e-08
sptr|Q7S412|Q7S412 Hypothetical protein. 55 6e-07
sptrnew|CAF05858|CAF05858 Related to alpha-L-arabinofuranosidase A. 55 6e-07
sptr|Q97DN6|Q97DN6 Probable alpha-arabinofuranosidase. 52 5e-06
sptr|Q8A359|Q8A359 Alpha-L-arabinofuranosidase A precursor. 49 4e-05
sptr|Q8G7R6|Q8G7R6 Similar to alpha-arabinofuranosidase I. 43 0.003
sptr|Q8G579|Q8G579 Similar to alpha-L-arabinofuranosidase A. 37 0.22
sptr|Q9NHX5|Q9NHX5 DESERT. 35 0.84
>sptr|Q9ATV8|Q9ATV8 Arabinoxylan arabinofuranohydrolase isoenzyme
AXAH-I.
Length = 658
Score = 291 bits (744), Expect = 7e-78
Identities = 137/164 (83%), Positives = 148/164 (90%)
Frame = -1
Query: 492 LLRFLFLLCIGCKCLASEVEATQTATLAVDASPQLARKIPDTLFGIFFEEINHAGAGGIW 313
+L FL L C+ CKCLASE E TQ A+L VD+SP LARKIPDTLFGIFFEEINHAGAGGIW
Sbjct: 13 ILCFLLLFCVSCKCLASEFEITQVASLGVDSSPHLARKIPDTLFGIFFEEINHAGAGGIW 72
Query: 312 AELVSNRGFEAGGLHTPSNIDPWSIIGDDSSVFVETDRTSCFSRNIVALRMEVLCDNCPT 133
AELVSNRGFEAGG HTPSNI+PWSIIGDDSS+FV TDRTSCFSRN VALRMEVLCDNCP
Sbjct: 73 AELVSNRGFEAGGPHTPSNINPWSIIGDDSSIFVGTDRTSCFSRNKVALRMEVLCDNCPV 132
Query: 132 GGVGVYNPGFWGMNIEEGKVYNLVMFVKSPETTDLTVSLTSSNG 1
GGVG+YNPGFWGMNIE+GK YNLVM VKSPET ++TVSLTSS+G
Sbjct: 133 GGVGIYNPGFWGMNIEDGKTYNLVMHVKSPETIEMTVSLTSSDG 176
>sptr|Q9ATV7|Q9ATV7 Arabinoxylan arabinofuranohydrolase isoenzyme
AXAH-II.
Length = 656
Score = 290 bits (742), Expect = 1e-77
Identities = 135/160 (84%), Positives = 147/160 (91%)
Frame = -1
Query: 480 LFLLCIGCKCLASEVEATQTATLAVDASPQLARKIPDTLFGIFFEEINHAGAGGIWAELV 301
L L C GCKC+ASE++ATQTATL VDAS QLARKIPDTLFG+FFEEINHAGAGGIWAELV
Sbjct: 15 LLLFCFGCKCIASELQATQTATLKVDASSQLARKIPDTLFGMFFEEINHAGAGGIWAELV 74
Query: 300 SNRGFEAGGLHTPSNIDPWSIIGDDSSVFVETDRTSCFSRNIVALRMEVLCDNCPTGGVG 121
SNRGFEAGGLHTPSNIDPWSIIGDDSS+FV TDRTSCFSRNI+ALRMEVLCD+CP GVG
Sbjct: 75 SNRGFEAGGLHTPSNIDPWSIIGDDSSIFVATDRTSCFSRNIIALRMEVLCDDCPASGVG 134
Query: 120 VYNPGFWGMNIEEGKVYNLVMFVKSPETTDLTVSLTSSNG 1
+YNPGFWGMNIE+GK YNLVM+VKS E +LTVSL SS+G
Sbjct: 135 IYNPGFWGMNIEDGKTYNLVMYVKSAEAAELTVSLASSDG 174
>sptr|Q7XWX2|Q7XWX2 OSJNBb0058J09.4 protein.
Length = 654
Score = 263 bits (673), Expect = 1e-69
Identities = 125/167 (74%), Positives = 143/167 (85%)
Frame = -1
Query: 501 RAVLLRFLFLLCIGCKCLASEVEATQTATLAVDASPQLARKIPDTLFGIFFEEINHAGAG 322
R ++ L LLC+ CKCL SEV TQ A L VDASPQ ARKIP+TLFGIFFEEINHAGAG
Sbjct: 8 RRIIFCVLLLLCVSCKCLTSEVNTTQLAVLKVDASPQHARKIPETLFGIFFEEINHAGAG 67
Query: 321 GIWAELVSNRGFEAGGLHTPSNIDPWSIIGDDSSVFVETDRTSCFSRNIVALRMEVLCDN 142
GIWAELVSNRGFEAGG +TPS+IDPWSIIG++S + V TDR+SCFSRNI+ALRMEVLC +
Sbjct: 68 GIWAELVSNRGFEAGGPNTPSSIDPWSIIGNESVISVATDRSSCFSRNIIALRMEVLCGD 127
Query: 141 CPTGGVGVYNPGFWGMNIEEGKVYNLVMFVKSPETTDLTVSLTSSNG 1
C GGVG+YNPGFWGMNIE+GK Y+LVM+ KS E T+LTVSLTSS+G
Sbjct: 128 CQAGGVGIYNPGFWGMNIEDGKNYSLVMYAKSLENTELTVSLTSSDG 174
>sptrnew|BAC99302|BAC99302 Alpha-L-arabinofuranosidase.
Length = 674
Score = 233 bits (595), Expect = 1e-60
Identities = 112/170 (65%), Positives = 137/170 (80%), Gaps = 5/170 (2%)
Frame = -1
Query: 495 VLLRFLFLLCIGCKCLASEVEATQTATLAVDASPQLARKIPDTLFGIFFEEINHAGAGGI 316
VLL LF L C+C A+ VEA QTA L V+AS AR+IPDTLFGIFFEEINHAGAGG+
Sbjct: 10 VLLLVLFGLSALCQCSATGVEANQTAVLLVNASEASARRIPDTLFGIFFEEINHAGAGGL 69
Query: 315 WAELVSNRGFEAGGLHTPSNIDPWSIIGDDSSVFVETDRTSCFSRNIVALRMEVLCDN-- 142
WAELV+NRGFE GG + PSNIDPWSIIGD+S V V TDR+SCF RN +A++++VLCD+
Sbjct: 70 WAELVNNRGFEGGGPNVPSNIDPWSIIGDESKVIVSTDRSSCFDRNKIAVQVQVLCDHTG 129
Query: 141 ---CPTGGVGVYNPGFWGMNIEEGKVYNLVMFVKSPETTDLTVSLTSSNG 1
CP GGVG+YNPGFWGMNIE+GK Y LV++V+S E+ +++V+LT SNG
Sbjct: 130 ANICPDGGVGIYNPGFWGMNIEQGKSYKLVLYVRSEESVNVSVALTGSNG 179
>sptr|Q944F3|Q944F3 Arabinosidase ARA-1.
Length = 674
Score = 233 bits (595), Expect = 1e-60
Identities = 112/170 (65%), Positives = 137/170 (80%), Gaps = 5/170 (2%)
Frame = -1
Query: 495 VLLRFLFLLCIGCKCLASEVEATQTATLAVDASPQLARKIPDTLFGIFFEEINHAGAGGI 316
VLL LF L C+C A+ VEA QTA L V+AS AR+IPDTLFGIFFEEINHAGAGG+
Sbjct: 10 VLLLVLFGLSALCQCSATGVEANQTAVLLVNASEASARRIPDTLFGIFFEEINHAGAGGL 69
Query: 315 WAELVSNRGFEAGGLHTPSNIDPWSIIGDDSSVFVETDRTSCFSRNIVALRMEVLCDN-- 142
WAELV+NRGFE GG + PSNIDPWSIIGD+S V V TDR+SCF RN +A++++VLCD+
Sbjct: 70 WAELVNNRGFEGGGPNVPSNIDPWSIIGDESKVIVSTDRSSCFDRNKIAVQVQVLCDHTG 129
Query: 141 ---CPTGGVGVYNPGFWGMNIEEGKVYNLVMFVKSPETTDLTVSLTSSNG 1
CP GGVG+YNPGFWGMNIE+GK Y LV++V+S E+ +++V+LT SNG
Sbjct: 130 ANICPDGGVGIYNPGFWGMNIEQGKSYKLVLYVRSEESVNVSVALTGSNG 179
>sptr|Q7X9G7|Q7X9G7 Alpha-L-arabinofuranosidase.
Length = 675
Score = 233 bits (593), Expect = 2e-60
Identities = 115/174 (66%), Positives = 138/174 (79%), Gaps = 5/174 (2%)
Frame = -1
Query: 507 APRAVLLRFLFLLCIGCKCLASEVEATQTATLAVDASPQLARKIPDTLFGIFFEEINHAG 328
+P VLL + LLC +C A V+A QTA L +DAS AR I DTLFGIFFEEINHAG
Sbjct: 6 SPHVVLLLHV-LLCSVYRCFAIGVDANQTANLLIDASQASARPISDTLFGIFFEEINHAG 64
Query: 327 AGGIWAELVSNRGFEAGGLHTPSNIDPWSIIGDDSSVFVETDRTSCFSRNIVALRMEVLC 148
AGG+WAELVSNRGFEAGG +TPSNIDPW+IIG++SS+ V TDR+SCF RN VALRMEVLC
Sbjct: 65 AGGVWAELVSNRGFEAGGPNTPSNIDPWAIIGNESSLIVSTDRSSCFDRNKVALRMEVLC 124
Query: 147 D-----NCPTGGVGVYNPGFWGMNIEEGKVYNLVMFVKSPETTDLTVSLTSSNG 1
D +CP GGVG+YNPGFWGMNIE+GK YN V++V+S + +++VSLT S+G
Sbjct: 125 DTQGANSCPAGGVGIYNPGFWGMNIEKGKTYNAVLYVRSSGSINVSVSLTGSDG 178
>sptrnew|BAC99303|BAC99303 Alpha-L-arabinofuranosidase.
Length = 674
Score = 231 bits (590), Expect = 5e-60
Identities = 114/174 (65%), Positives = 138/174 (79%), Gaps = 5/174 (2%)
Frame = -1
Query: 507 APRAVLLRFLFLLCIGCKCLASEVEATQTATLAVDASPQLARKIPDTLFGIFFEEINHAG 328
+P VLL ++ LLC +C A V+A QTA L +DAS AR I DTLFGIFFEEINHAG
Sbjct: 6 SPHIVLLLYV-LLCSVYRCFAIGVDANQTANLLIDASQASARPISDTLFGIFFEEINHAG 64
Query: 327 AGGIWAELVSNRGFEAGGLHTPSNIDPWSIIGDDSSVFVETDRTSCFSRNIVALRMEVLC 148
AGG+WAELVSNRGFEAGG +TPSNIDPW+IIG++S + V TDR+SCF RN VALRMEVLC
Sbjct: 65 AGGVWAELVSNRGFEAGGPNTPSNIDPWAIIGNESFLIVSTDRSSCFDRNKVALRMEVLC 124
Query: 147 D-----NCPTGGVGVYNPGFWGMNIEEGKVYNLVMFVKSPETTDLTVSLTSSNG 1
D +CP GGVG+YNPGFWGMNIE+GK YN V++V+S + +++VSLT S+G
Sbjct: 125 DTQGANSCPAGGVGIYNPGFWGMNIEKGKTYNAVLYVRSSGSINVSVSLTGSDG 178
>sptr|Q8LIG5|Q8LIG5 Putative arabinoxylan narabinofuranohydrolase
isoenzyme AXAH-I.
Length = 664
Score = 225 bits (574), Expect = 3e-58
Identities = 116/182 (63%), Positives = 136/182 (74%), Gaps = 5/182 (2%)
Frame = -1
Query: 531 MGITSNDAAPRAVLLRFLFLLCIGCKCLASEVEATQTATLAVDASPQLARKIPDTLFGIF 352
M + DAA LR L L+ C ++ A QT L VDASPQ ARKIPD +FGIF
Sbjct: 1 MAVQRTDAAS---WLRELVLVYAMCWTVSLGFVAGQTGQLNVDASPQNARKIPDKMFGIF 57
Query: 351 FEEINHAGAGGIWAELVSNRGFEAGGLHTPSNIDPWSIIGDDSSVFVETDRTSCFSRNIV 172
FEEINHAGAGG+WAELVSNRGFEAGG +TPSNIDPW IIG++SS+ V TDRTSCF +N V
Sbjct: 58 FEEINHAGAGGLWAELVSNRGFEAGGPNTPSNIDPWLIIGNESSIIVGTDRTSCFEKNPV 117
Query: 171 ALRMEVLCD-----NCPTGGVGVYNPGFWGMNIEEGKVYNLVMFVKSPETTDLTVSLTSS 7
ALRMEVLCD NCP+GGVGVYNPG+WGMNIE +VY + + ++S + LTVSLTSS
Sbjct: 118 ALRMEVLCDSKGTNNCPSGGVGVYNPGYWGMNIERRRVYKVGLHIRSSDAVSLTVSLTSS 177
Query: 6 NG 1
+G
Sbjct: 178 DG 179
>sptr|Q84RC5|Q84RC5 Alpha-L-arabinofuranosidase.
Length = 678
Score = 216 bits (551), Expect = 2e-55
Identities = 107/169 (63%), Positives = 130/169 (76%), Gaps = 5/169 (2%)
Frame = -1
Query: 492 LLRFLFLLCI---GCKCLASEVEATQTATLAVDASPQLARKIPDTLFGIFFEEINHAGAG 322
+L FL C + + ++ + TL VDAS R IP+TLFGIFFEEINHAGAG
Sbjct: 14 VLSFLLGSCFVYQSLRVVDAQEDPKPAVTLQVDASNGGGRPIPETLFGIFFEEINHAGAG 73
Query: 321 GIWAELVSNRGFEAGGLHTPSNIDPWSIIGDDSSVFVETDRTSCFSRNIVALRMEVLCDN 142
G+WAELVSNRGFEAGG +TPSNI PWSI+GD SS++V TDR+SCF RN +ALRM+VLCD+
Sbjct: 74 GLWAELVSNRGFEAGGQNTPSNIWPWSIVGDHSSIYVATDRSSCFERNKIALRMDVLCDS 133
Query: 141 --CPTGGVGVYNPGFWGMNIEEGKVYNLVMFVKSPETTDLTVSLTSSNG 1
CP+GGVGVYNPG+WGMNIEEGK Y + ++V+S DL+VSLTSSNG
Sbjct: 134 KGCPSGGVGVYNPGYWGMNIEEGKKYKVALYVRSTGDIDLSVSLTSSNG 182
>sptr|Q94JT7|Q94JT7 AT3g10740/T7M13_18.
Length = 678
Score = 216 bits (551), Expect = 2e-55
Identities = 107/169 (63%), Positives = 130/169 (76%), Gaps = 5/169 (2%)
Frame = -1
Query: 492 LLRFLFLLCI---GCKCLASEVEATQTATLAVDASPQLARKIPDTLFGIFFEEINHAGAG 322
+L FL C + + ++ + TL VDAS R IP+TLFGIFFEEINHAGAG
Sbjct: 14 VLSFLLGSCFVYQSLRVVDAQEDPKPAVTLQVDASNGGGRPIPETLFGIFFEEINHAGAG 73
Query: 321 GIWAELVSNRGFEAGGLHTPSNIDPWSIIGDDSSVFVETDRTSCFSRNIVALRMEVLCDN 142
G+WAELVSNRGFEAGG +TPSNI PWSI+GD SS++V TDR+SCF RN +ALRM+VLCD+
Sbjct: 74 GLWAELVSNRGFEAGGQNTPSNIWPWSIVGDHSSIYVATDRSSCFERNKIALRMDVLCDS 133
Query: 141 --CPTGGVGVYNPGFWGMNIEEGKVYNLVMFVKSPETTDLTVSLTSSNG 1
CP+GGVGVYNPG+WGMNIEEGK Y + ++V+S DL+VSLTSSNG
Sbjct: 134 KGCPSGGVGVYNPGYWGMNIEEGKKYKVALYVRSTGDIDLSVSLTSSNG 182
>sptr|Q9SG80|Q9SG80 Putative alpha-L-arabinofuranosidase.
Length = 678
Score = 216 bits (551), Expect = 2e-55
Identities = 107/169 (63%), Positives = 130/169 (76%), Gaps = 5/169 (2%)
Frame = -1
Query: 492 LLRFLFLLCI---GCKCLASEVEATQTATLAVDASPQLARKIPDTLFGIFFEEINHAGAG 322
+L FL C + + ++ + TL VDAS R IP+TLFGIFFEEINHAGAG
Sbjct: 14 VLSFLLGSCFVYQSLRVVDAQEDPKPAVTLQVDASNGGGRPIPETLFGIFFEEINHAGAG 73
Query: 321 GIWAELVSNRGFEAGGLHTPSNIDPWSIIGDDSSVFVETDRTSCFSRNIVALRMEVLCDN 142
G+WAELVSNRGFEAGG +TPSNI PWSI+GD SS++V TDR+SCF RN +ALRM+VLCD+
Sbjct: 74 GLWAELVSNRGFEAGGQNTPSNIWPWSIVGDHSSIYVATDRSSCFERNKIALRMDVLCDS 133
Query: 141 --CPTGGVGVYNPGFWGMNIEEGKVYNLVMFVKSPETTDLTVSLTSSNG 1
CP+GGVGVYNPG+WGMNIEEGK Y + ++V+S DL+VSLTSSNG
Sbjct: 134 KGCPSGGVGVYNPGYWGMNIEEGKKYKVALYVRSTGDIDLSVSLTSSNG 182
>sptr|Q8VZR2|Q8VZR2 Putative arabinosidase
(Alpha-L-arabinofuranosidase) (Hypothetical protein
At5g26120).
Length = 674
Score = 213 bits (543), Expect = 1e-54
Identities = 106/150 (70%), Positives = 123/150 (82%), Gaps = 4/150 (2%)
Frame = -1
Query: 438 VEATQTA--TLAVDASPQLARKIPDTLFGIFFEEINHAGAGGIWAELVSNRGFEAGGLHT 265
V+A + A TL VDAS R IP+TLFGIFFEEINHAGAGG+WAELVSNRGFEAGG
Sbjct: 32 VDAQEDAIVTLQVDASNVTRRPIPETLFGIFFEEINHAGAGGLWAELVSNRGFEAGGQII 91
Query: 264 PSNIDPWSIIGDDSSVFVETDRTSCFSRNIVALRMEVLCD--NCPTGGVGVYNPGFWGMN 91
PSNI PWSIIGD+SS++V TDR+SCF RN +ALRMEVLCD +CP GGVGVYNPG+WGMN
Sbjct: 92 PSNIWPWSIIGDESSIYVVTDRSSCFERNKIALRMEVLCDSNSCPLGGVGVYNPGYWGMN 151
Query: 90 IEEGKVYNLVMFVKSPETTDLTVSLTSSNG 1
IEEGK Y +V++V+S D++VS TSSNG
Sbjct: 152 IEEGKKYKVVLYVRSTGDIDVSVSFTSSNG 181
>sptrnew|AAR27067|AAR27067 A-arabinofuranosidase 1 (Fragment).
Length = 272
Score = 99.8 bits (247), Expect = 3e-20
Identities = 43/76 (56%), Positives = 60/76 (78%), Gaps = 5/76 (6%)
Frame = -1
Query: 213 VETDRTSCFSRNIVALRMEVLCDN-----CPTGGVGVYNPGFWGMNIEEGKVYNLVMFVK 49
V TDR+SCF RN +AL+MEVLCD+ CP GVG+YNPG+WGMNIE+GK Y ++++V+
Sbjct: 1 VSTDRSSCFERNKIALKMEVLCDSKGPNICPPEGVGIYNPGYWGMNIEQGKSYKVILYVR 60
Query: 48 SPETTDLTVSLTSSNG 1
S + +++V+LT SNG
Sbjct: 61 SDDAINVSVALTGSNG 76
>sptr|Q9XH04|Q9XH04 T1N24.13 protein.
Length = 521
Score = 83.6 bits (205), Expect = 2e-15
Identities = 38/56 (67%), Positives = 46/56 (82%), Gaps = 2/56 (3%)
Frame = -1
Query: 162 MEVLCDN--CPTGGVGVYNPGFWGMNIEEGKVYNLVMFVKSPETTDLTVSLTSSNG 1
MEVLCD+ CP GGVGVYNPG+WGMNIEEGK Y +V++V+S D++VS TSSNG
Sbjct: 1 MEVLCDSNSCPLGGVGVYNPGYWGMNIEEGKKYKVVLYVRSTGDIDVSVSFTSSNG 56
>sptr|Q828B3|Q828B3 Putative secreted arabinosidase.
Length = 710
Score = 69.7 bits (169), Expect = 3e-11
Identities = 59/189 (31%), Positives = 94/189 (49%), Gaps = 3/189 (1%)
Frame = -1
Query: 558 PTRVDKWKRMGITSNDAAPRAVLLRFLFLLCIGCKCLASEVEATQTATLAVDASPQLARK 379
PTR +W R+G+T+ L + +G A + T A +AVD A K
Sbjct: 3 PTRT-RW-RLGLTA----------AALLVASVGVPAPAHAEDITDYA-IAVDPKGSGA-K 48
Query: 378 IPDTLFGIFFEEINHAGAGGIWAELVSNRGFEAGGLHTPS--NIDPWSIIGDDSSVFVET 205
I DT++G+FFE+IN A GG++AELV NR FE S + W+ G + V +
Sbjct: 49 IDDTMYGVFFEDINRAADGGLYAELVQNRSFEYATADNTSYTPLTSWNTSG-TADVVSDD 107
Query: 204 DRTSCFSRNIVALRMEVLCDNCPTGGVGVYNPGF-WGMNIEEGKVYNLVMFVKSPETTDL 28
R + +R+ +AL G V N G+ G+ +E GKVY+ ++ ++ + L
Sbjct: 108 GRLNARNRSYLAL----------GGDSSVTNSGYNTGIAVESGKVYDFSVWARADQADPL 157
Query: 27 TVSLTSSNG 1
+V+L ++G
Sbjct: 158 SVTLHDTDG 166
>sptr|O88043|O88043 Putative secreted arabinosidase.
Length = 824
Score = 66.2 bits (160), Expect = 3e-10
Identities = 48/151 (31%), Positives = 78/151 (51%), Gaps = 3/151 (1%)
Frame = -1
Query: 444 SEVEATQTATLAVDASPQLARKIPDTLFGIFFEEINHAGAGGIWAELVSNRGFEAGGLHT 265
+ EA T+ VD S + I DT++G+F+E+IN A GG++AELV NR FE
Sbjct: 26 AHAEAVADYTITVDPSDR-GPAIDDTMYGVFYEDINRAADGGLYAELVQNRSFEYSTADN 84
Query: 264 PS--NIDPWSIIGDDSSVFVETDRTSCFSRNIVALRMEVLCDNCPTGGVGVYNPGF-WGM 94
S + W+ G + V + R + +RN ++L G V N G+ G+
Sbjct: 85 ASYTPLTAWAADG-TAEVVNDGGRLNERNRNYLSL----------AAGSTVTNAGYNTGI 133
Query: 93 NIEEGKVYNLVMFVKSPETTDLTVSLTSSNG 1
+E+G+ Y+ ++ ++ T LTV+LT + G
Sbjct: 134 RVEKGERYDFSVWSRAGHGTTLTVTLTDAAG 164
>sptr|Q8NK90|Q8NK90 Alpha-L-arabinofuranosidase A (EC 3.2.1.55).
Length = 628
Score = 66.2 bits (160), Expect = 3e-10
Identities = 41/138 (29%), Positives = 73/138 (52%), Gaps = 2/138 (1%)
Frame = -1
Query: 414 LAVDASPQLARKIPDTLFGIFFEEINHAGAGGIWAELVSNRGFEAGGLHTPSNIDPWSII 235
+++ S Q L+G FE+INH+G GGI+ +L+ N G + T N+ W+ +
Sbjct: 26 VSLKVSTQGGNSSSPILYGFMFEDINHSGDGGIYGQLLQNPGLQG----TTPNLTAWAAV 81
Query: 234 GDDSSVFVETDR--TSCFSRNIVALRMEVLCDNCPTGGVGVYNPGFWGMNIEEGKVYNLV 61
G D+++ ++ D TS I +++V D TG VG+ N G+WG+ + +G +
Sbjct: 82 G-DATIAIDGDSPLTSAIPSTI---KLDVADD--ATGAVGLTNEGYWGIPV-DGSEFQSS 134
Query: 60 MFVKSPETTDLTVSLTSS 7
++K + D+TV L +
Sbjct: 135 FWIKGEYSGDITVRLVGN 152
>sptr|Q96X54|Q96X54 Alpha-L-arabinofuranosidase A (EC 3.2.1.55).
Length = 628
Score = 66.2 bits (160), Expect = 3e-10
Identities = 41/138 (29%), Positives = 73/138 (52%), Gaps = 2/138 (1%)
Frame = -1
Query: 414 LAVDASPQLARKIPDTLFGIFFEEINHAGAGGIWAELVSNRGFEAGGLHTPSNIDPWSII 235
+++ S Q L+G FE+INH+G GGI+ +L+ N G + T N+ W+ +
Sbjct: 26 VSLKVSTQGGNSSSPILYGFMFEDINHSGDGGIYGQLLQNPGLQG----TTPNLTAWAAV 81
Query: 234 GDDSSVFVETDR--TSCFSRNIVALRMEVLCDNCPTGGVGVYNPGFWGMNIEEGKVYNLV 61
G D+++ ++ D TS I +++V D TG VG+ N G+WG+ + +G +
Sbjct: 82 G-DATIAIDGDSPLTSAIPSTI---KLDVADD--ATGAVGLTNEGYWGIPV-DGSEFQSS 134
Query: 60 MFVKSPETTDLTVSLTSS 7
++K + D+TV L +
Sbjct: 135 FWIKGDYSGDITVRLVGN 152
>sw|P42254|ABFA_ASPNG Alpha-L-arabinofuranosidase A precursor (EC
3.2.1.55) (Arabinosidase A) (ABF A).
Length = 628
Score = 65.1 bits (157), Expect = 8e-10
Identities = 39/138 (28%), Positives = 73/138 (52%), Gaps = 2/138 (1%)
Frame = -1
Query: 414 LAVDASPQLARKIPDTLFGIFFEEINHAGAGGIWAELVSNRGFEAGGLHTPSNIDPWSII 235
+++ S Q L+G FE+INH+G GGI+ +++ N G + T N+ W+ +
Sbjct: 26 ISLKVSTQGGNSSSPILYGFMFEDINHSGDGGIYGQMLQNPGLQG----TAPNLTAWAAV 81
Query: 234 GDDSSVFVETDR--TSCFSRNIVALRMEVLCDNCPTGGVGVYNPGFWGMNIEEGKVYNLV 61
G D+++ ++ D TS I ++ + D TG VG+ N G+WG+ + +G ++
Sbjct: 82 G-DATIAIDGDSPLTSAIPSTI---KLNIADD--ATGAVGLTNEGYWGIPV-DGSEFHSS 134
Query: 60 MFVKSPETTDLTVSLTSS 7
++K + D+TV L +
Sbjct: 135 FWIKGDYSGDITVRLVGN 152
>sptr|Q8A1K4|Q8A1K4 Alpha-L-arabinofuranosidase A precursor.
Length = 825
Score = 64.3 bits (155), Expect = 1e-09
Identities = 45/144 (31%), Positives = 74/144 (51%), Gaps = 8/144 (5%)
Frame = -1
Query: 408 VDASPQLARKIPDTLFGIFFEEINHAGAGGIWAELVSNRGFE-----AGGLHTPSNIDPW 244
V A P+ ++I D L GIFFE+IN++ GG++AEL+ NR FE G ++ W
Sbjct: 195 VTAQPEETKEISDLLMGIFFEDINYSADGGLYAELIQNRDFEYEPSDREGDKNWNSTHSW 254
Query: 243 SIIGDDSSVFVETDRTSCFSRNIVALRMEVLCDNCPTGGVGVYNPGFWGMNIEEGKVYNL 64
+ G++++ + T + A VL N P G + N GF G+ ++ G+ Y+
Sbjct: 255 KLEGENATFTISTSDPIHPNNPHYA----VLKTNQP--GAALTNTGFDGIALKAGEKYDF 308
Query: 63 VMFVKSPE---TTDLTVSLTSSNG 1
+F + PE + L V L ++G
Sbjct: 309 SLFARIPEGSKSGKLLVRLVDADG 332
>sw|P82593|ABF1_STRCX Alpha-L-arabinofuranosidase I precursor (EC
3.2.1.55) (Arabinosidase I) (Alpha-L-AFase I).
Length = 825
Score = 63.2 bits (152), Expect = 3e-09
Identities = 43/131 (32%), Positives = 69/131 (52%), Gaps = 5/131 (3%)
Frame = -1
Query: 378 IPDTLFGIFFEEINHAGAGGIWAELVSNRGFE----AGGLHTPSNIDPWSIIGDDSSVFV 211
I DT++G+FFE+IN A GG++AELV NR FE +TP + W I+ V
Sbjct: 48 IDDTMYGVFFEDINRAADGGLYAELVQNRSFEYSTDDNRSYTP--LTSW-IVDGTGEVVN 104
Query: 210 ETDRTSCFSRNIVALRMEVLCDNCPTGGVGVYNPGF-WGMNIEEGKVYNLVMFVKSPETT 34
+ R + +RN ++L G V N G+ G+ +E+GK Y+ ++ ++ +
Sbjct: 105 DAGRLNERNRNYLSL----------GAGSSVTNAGYNTGIRVEQGKRYDFSVWARAGSAS 154
Query: 33 DLTVSLTSSNG 1
LTV+L + G
Sbjct: 155 TLTVALKDAAG 165
>sptr|Q59218|Q59218 Arabinosidase (EC 3.2.1.55).
Length = 660
Score = 58.9 bits (141), Expect = 5e-08
Identities = 39/129 (30%), Positives = 64/129 (49%)
Frame = -1
Query: 426 QTATLAVDASPQLARKIPDTLFGIFFEEINHAGAGGIWAELVSNRGFEAGGLHTPSNIDP 247
QT L + +L +I T++G+FFE+IN+A GG++AELV NR FE P ++
Sbjct: 22 QTNELVIQTK-KLGAEIQPTMYGLFFEDINYAADGGLYAELVKNRSFE-----FPQHLMG 75
Query: 246 WSIIGDDSSVFVETDRTSCFSRNIVALRMEVLCDNCPTGGVGVYNPGFWGMNIEEGKVYN 67
W G V F RN +R+ G+ N GF+G+ +++G+ Y
Sbjct: 76 WKTYGK-----VSLMNDGPFERNPHYVRLS--DPGHAHKHTGLDNEGFFGIGVKKGEEYR 128
Query: 66 LVMFVKSPE 40
++ + P+
Sbjct: 129 FSVWARLPQ 137
>sptr|Q8AAU6|Q8AAU6 Alpha-L-arabinofuranosidase A precursor.
Length = 660
Score = 58.5 bits (140), Expect = 7e-08
Identities = 37/117 (31%), Positives = 62/117 (52%)
Frame = -1
Query: 393 QLARKIPDTLFGIFFEEINHAGAGGIWAELVSNRGFEAGGLHTPSNIDPWSIIGDDSSVF 214
+L +I T++G+FFE+IN+A GG++AELV NR FE P + W G V
Sbjct: 32 KLGAEIQPTMYGLFFEDINYAADGGLYAELVKNRSFE-----FPQRLMGWKTYG---KVT 83
Query: 213 VETDRTSCFSRNIVALRMEVLCDNCPTGGVGVYNPGFWGMNIEEGKVYNLVMFVKSP 43
++ D F RN +R++ G+ N GF+G+ +++G+ Y ++ + P
Sbjct: 84 LQDD--GPFERNPHYVRLD--NPGHAHKHTGLDNEGFFGIGVKQGEEYRFSVWARLP 136
>sptr|Q7S412|Q7S412 Hypothetical protein.
Length = 667
Score = 55.5 bits (132), Expect = 6e-07
Identities = 37/122 (30%), Positives = 61/122 (50%), Gaps = 2/122 (1%)
Frame = -1
Query: 366 LFGIFFEEINHAGAGGIWAELVSNRGFEAGG-LHTPSNIDPWSIIGDDSSVFVETDRTSC 190
++G+ E+IN++G GGI+AEL+ NR F++G P +D WS + + + + S
Sbjct: 45 MYGLMHEDINNSGDGGIYAELIRNRAFQSGDESRYPVTLDGWSAV---NGAKLSLKKLSQ 101
Query: 189 FSRNIVALRMEVLCDNCPTGGVGVYNPGFWGMNIEEGKVYNLVMFVKSP-ETTDLTVSLT 13
+ M V + VG N G+WGM+++ K Y +VK + T SL
Sbjct: 102 PLSAALPYSMNVAVPSSSNKTVGFANAGYWGMDVKVQK-YTGSFYVKGAYQGKSFTASLQ 160
Query: 12 SS 7
S+
Sbjct: 161 SA 162
>sptrnew|CAF05858|CAF05858 Related to alpha-L-arabinofuranosidase A.
Length = 656
Score = 55.5 bits (132), Expect = 6e-07
Identities = 37/122 (30%), Positives = 61/122 (50%), Gaps = 2/122 (1%)
Frame = -1
Query: 366 LFGIFFEEINHAGAGGIWAELVSNRGFEAGG-LHTPSNIDPWSIIGDDSSVFVETDRTSC 190
++G+ E+IN++G GGI+AEL+ NR F++G P +D WS + + + + S
Sbjct: 45 MYGLMHEDINNSGDGGIYAELIRNRAFQSGDESRYPVTLDGWSAV---NGAKLSLKKLSQ 101
Query: 189 FSRNIVALRMEVLCDNCPTGGVGVYNPGFWGMNIEEGKVYNLVMFVKSP-ETTDLTVSLT 13
+ M V + VG N G+WGM+++ K Y +VK + T SL
Sbjct: 102 PLSAALPYSMNVAVPSSSNKTVGFANAGYWGMDVKVQK-YTGSFYVKGAYQGKSFTASLQ 160
Query: 12 SS 7
S+
Sbjct: 161 SA 162
>sptr|Q97DN6|Q97DN6 Probable alpha-arabinofuranosidase.
Length = 835
Score = 52.4 bits (124), Expect = 5e-06
Identities = 32/127 (25%), Positives = 60/127 (47%), Gaps = 3/127 (2%)
Frame = -1
Query: 372 DTLFGIFFEEINHAGAGGIWAELVSNRGFEAGGLHTPSNIDPWSIIGDDSSVFVETDRTS 193
D L G+FFE+INH GG+++EL+ N+ FE ++ W++ D + +
Sbjct: 52 DMLTGLFFEDINHGADGGLYSELLQNQSFE-----FKDSLSSWTV---DKTGSTSSTAEV 103
Query: 192 CFSRNIVALRMEVLCDNCP--TGGVGVYNPGFWGMNIEEGKVYNLVMFVKS-PETTDLTV 22
C S+ + + L NCP + + N G+ G+ + Y+ + ++ + +T+
Sbjct: 104 CTSKPLNSNNTHYLELNCPDNNSSLKLVNSGYKGITVNNNAKYDFYFWARNVGKGNKVTI 163
Query: 21 SLTSSNG 1
L NG
Sbjct: 164 QLEDENG 170
>sptr|Q8A359|Q8A359 Alpha-L-arabinofuranosidase A precursor.
Length = 709
Score = 49.3 bits (116), Expect = 4e-05
Identities = 46/167 (27%), Positives = 72/167 (43%), Gaps = 28/167 (16%)
Frame = -1
Query: 444 SEVEATQTATLAVDASPQLARKIPDTLFGIFFEEINHAGAGGIWAELVSNRGFE------ 283
S+V T L +D Q RK+ +G +EEI G G + AELV NR FE
Sbjct: 53 SQVNDTHEGVLHIDK--QKTRKVSRVQYGFHYEEIGMIGEGALHAELVRNRSFEEATPPA 110
Query: 282 ----AGGLH--TPS---------NIDPWSIIG------DDSSVFV-ETDRTSCFSRNIVA 169
GL+ P+ ++DP +IG + +F+ T+ N +
Sbjct: 111 DLAVKNGLYQNVPNPRGKNKDVFHVDP--LIGWNTYPLSYTPIFISRTEENPLNKENKYS 168
Query: 168 LRMEVLCDNCPTGGVGVYNPGFWGMNIEEGKVYNLVMFVKSPETTDL 28
+ + V D + N G++GMN+ + Y+L M++KS T L
Sbjct: 169 MLVNVTEDIANNPEAMILNRGYYGMNLRKEVSYHLSMYIKSKNYTAL 215
>sptr|Q8G7R6|Q8G7R6 Similar to alpha-arabinofuranosidase I.
Length = 823
Score = 43.1 bits (100), Expect = 0.003
Identities = 22/49 (44%), Positives = 30/49 (61%), Gaps = 2/49 (4%)
Frame = -1
Query: 384 RKIPDTLFGIFFEEINHAGAGGIWAELVSNRGFEAGGLHTP--SNIDPW 244
RKI L+G+FFE+I+++G GG+ +ELV N FE P SN W
Sbjct: 17 RKISTDLWGVFFEDISYSGDGGLNSELVQNGAFEYNRADKPEWSNYTAW 65
>sptr|Q8G579|Q8G579 Similar to alpha-L-arabinofuranosidase A.
Length = 769
Score = 37.0 bits (84), Expect = 0.22
Identities = 14/28 (50%), Positives = 21/28 (75%)
Frame = -1
Query: 366 LFGIFFEEINHAGAGGIWAELVSNRGFE 283
L+G+FFE+INH GG+ A +V+N F+
Sbjct: 35 LYGVFFEDINHGADGGLNANMVNNYSFD 62
>sptr|Q9NHX5|Q9NHX5 DESERT.
Length = 1178
Score = 35.0 bits (79), Expect = 0.84
Identities = 24/94 (25%), Positives = 46/94 (48%), Gaps = 1/94 (1%)
Frame = +2
Query: 341 ISSKKIPNSVSGIFRASCGEASTARVAVWVASTSDA-RHLQPMQSRNKKRRRTARGAASL 517
+S+ K+P S+ SC E +T + AS SD+ + ++ ++ + KK+ T G A
Sbjct: 545 LSNGKVPGSLKPKSHKSCSETTTGNSSSATASASDSEQRVKSVKKQAKKKTTTKEGGAGK 604
Query: 518 LVIPIRFHLSTLVGPACLPCMHASTIRRKTRAPV 619
+ + + +T PA + + A+ R K + V
Sbjct: 605 VANANKTNSTTESPPAVVDVVPATQTRLKKLSAV 638
Database: /db/uniprot/tmp/swall
Posted date: Mar 5, 2004 7:45 PM
Number of letters in database: 442,889,342
Number of sequences in database: 1,395,590
Lambda K H
0.318 0.135 0.401
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 559,828,496
Number of Sequences: 1395590
Number of extensions: 12161152
Number of successful extensions: 37685
Number of sequences better than 10.0: 30
Number of HSP's better than 10.0 without gapping: 36249
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 37641
length of database: 442,889,342
effective HSP length: 118
effective length of database: 278,209,722
effective search space used: 26708133312
frameshift window, decay const: 50, 0.1
T: 12
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)

