BLASTX 2.2.6 [Apr-09-2003]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= 3276099.2.1
(545 letters)
Database: /db/trembl-ebi/tmp/swall
1,272,877 sequences; 406,886,216 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
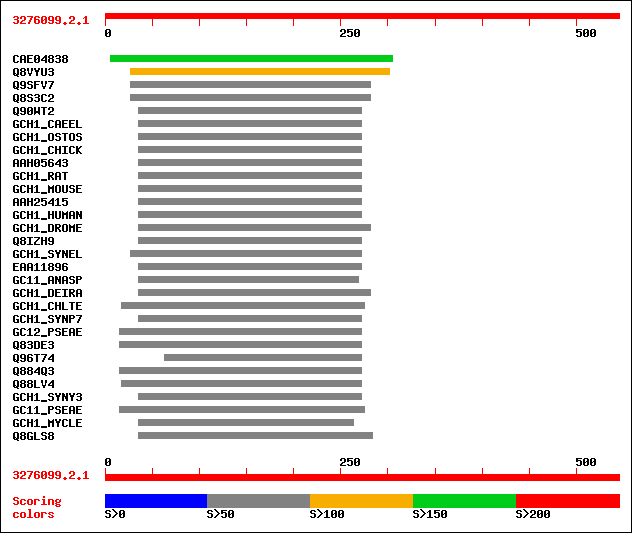
sptrnew|CAE04838|CAE04838 OSJNBa0084K01.10 protein. 173 1e-42
sptr|Q8VYU3|Q8VYU3 GTP cyclohydrolase I (EC 3.5.4.16). 108 4e-23
sptr|Q9SFV7|Q9SFV7 GTP cyclohydrolase I (At3g07270). 99 3e-20
sptr|Q8S3C2|Q8S3C2 GTP cyclohydrolase I. 99 3e-20
sptr|Q90WT2|Q90WT2 GTP cyclohydrolase I. 72 5e-12
sw|Q19980|GCH1_CAEEL Probable GTP cyclohydrolase I (EC 3.5.4.16)... 72 5e-12
sw|O61573|GCH1_OSTOS GTP cyclohydrolase I (EC 3.5.4.16) (GTP-CH-I). 70 1e-11
sw|P50141|GCH1_CHICK GTP cyclohydrolase I (EC 3.5.4.16) (GTP-CH-I). 70 2e-11
sptrnew|AAH05643|AAH05643 GTP cyclohydrolase 1. 69 4e-11
sw|P22288|GCH1_RAT GTP cyclohydrolase I precursor (EC 3.5.4.16) ... 69 4e-11
sw|Q05915|GCH1_MOUSE GTP cyclohydrolase I precursor (EC 3.5.4.16... 69 4e-11
sptrnew|AAH25415|AAH25415 GTP cyclohydrolase 1 (Dopa-responsive ... 69 4e-11
sw|P30793|GCH1_HUMAN GTP cyclohydrolase I (EC 3.5.4.16) (GTP-CH-I). 69 4e-11
sw|P48596|GCH1_DROME GTP cyclohydrolase I (EC 3.5.4.16) (GTP-CH-... 69 4e-11
sptr|Q8IZH9|Q8IZH9 GTP cyclohydrolase I type IV (EC 3.5.4.16) (F... 69 4e-11
sw|Q8DJB8|GCH1_SYNEL GTP cyclohydrolase I (EC 3.5.4.16) (GTP-CH-I). 67 1e-10
sptrnew|EAA11896|EAA11896 AgCP6428 (Fragment). 67 1e-10
sw|Q8YLL1|GC11_ANASP GTP cyclohydrolase I 1 (EC 3.5.4.16) (GTP-C... 67 2e-10
sw|Q9RYB4|GCH1_DEIRA GTP cyclohydrolase I (EC 3.5.4.16) (GTP-CH-I). 67 2e-10
sw|Q8KEA8|GCH1_CHLTE GTP cyclohydrolase I (EC 3.5.4.16) (GTP-CH-I). 67 2e-10
sw|Q54769|GCH1_SYNP7 GTP cyclohydrolase I (EC 3.5.4.16) (GTP-CH-I). 66 2e-10
sw|Q9I351|GC12_PSEAE GTP cyclohydrolase I 2 (EC 3.5.4.16) (GTP-C... 66 2e-10
sptr|Q83DE3|Q83DE3 GTP cyclohydrolase I. 66 2e-10
sptr|Q96T74|Q96T74 GTP cyclohydrolase I (Fragment). 66 3e-10
sptr|Q884Q3|Q884Q3 GTP cyclohydrolase I. 65 4e-10
sptr|Q88LV4|Q88LV4 GTP cyclohydrolase I. 65 4e-10
sw|Q55759|GCH1_SYNY3 GTP cyclohydrolase I (EC 3.5.4.16) (GTP-CH-I). 65 4e-10
sw|Q9HYG8|GC11_PSEAE GTP cyclohydrolase I 1 (EC 3.5.4.16) (GTP-C... 65 5e-10
sw|O69531|GCH1_MYCLE GTP cyclohydrolase I (EC 3.5.4.16) (GTP-CH-I). 64 1e-09
sptr|Q8GLS8|Q8GLS8 GTP cyclohydrolase I (EC 3.5.4.16). 64 1e-09
>sptrnew|CAE04838|CAE04838 OSJNBa0084K01.10 protein.
Length = 451
Score = 173 bits (438), Expect = 1e-42
Identities = 88/100 (88%), Positives = 92/100 (92%)
Frame = +3
Query: 6 DNGSGEGIDRSHFQALVHFYGCKLQVQERMTRQIAEAVYSVSHNGAMVVVEANHICMISR 185
D G GE IDRSHFQALVHFYGCKLQVQERMTRQIAEAVYSVSH GA+VVVEANHICMISR
Sbjct: 352 DGGDGEVIDRSHFQALVHFYGCKLQVQERMTRQIAEAVYSVSHCGAIVVVEANHICMISR 411
Query: 186 GIEKIRSNTATIAVLGQFLADPSAKACFLQNVLDSVGSAV 305
GIEKIRS+TATIAVLGQFL DPSAKA FLQNV+D+ G AV
Sbjct: 412 GIEKIRSSTATIAVLGQFLTDPSAKARFLQNVVDTTGLAV 451
>sptr|Q8VYU3|Q8VYU3 GTP cyclohydrolase I (EC 3.5.4.16).
Length = 456
Score = 108 bits (270), Expect = 4e-23
Identities = 60/92 (65%), Positives = 69/92 (75%)
Frame = +3
Query: 27 IDRSHFQALVHFYGCKLQVQERMTRQIAEAVYSVSHNGAMVVVEANHICMISRGIEKIRS 206
+ R Q++VHFYG KLQVQER+TRQIAE V S +VVVEANH CMISRGIEK S
Sbjct: 364 VGRPLVQSVVHFYGFKLQVQERVTRQIAETVSSFLGEDIIVVVEANHTCMISRGIEKFGS 423
Query: 207 NTATIAVLGQFLADPSAKACFLQNVLDSVGSA 302
NTAT AVLG+F DP A+A FLQ++ DS GSA
Sbjct: 424 NTATFAVLGRFSTDPVARAKFLQSLPDS-GSA 454
>sptr|Q9SFV7|Q9SFV7 GTP cyclohydrolase I (At3g07270).
Length = 466
Score = 99.0 bits (245), Expect = 3e-20
Identities = 51/85 (60%), Positives = 62/85 (72%)
Frame = +3
Query: 27 IDRSHFQALVHFYGCKLQVQERMTRQIAEAVYSVSHNGAMVVVEANHICMISRGIEKIRS 206
+ S +A+VHFYG KLQVQERMTRQIAE + + +VV EA H CMISRGIEK S
Sbjct: 368 VGSSLMKAIVHFYGFKLQVQERMTRQIAETLSPLVGGDVIVVAEAGHTCMISRGIEKFGS 427
Query: 207 NTATIAVLGQFLADPSAKACFLQNV 281
+TATIAVLG+F +D SA+A FL +
Sbjct: 428 STATIAVLGRFSSDNSARAMFLDKI 452
>sptr|Q8S3C2|Q8S3C2 GTP cyclohydrolase I.
Length = 466
Score = 99.0 bits (245), Expect = 3e-20
Identities = 51/85 (60%), Positives = 62/85 (72%)
Frame = +3
Query: 27 IDRSHFQALVHFYGCKLQVQERMTRQIAEAVYSVSHNGAMVVVEANHICMISRGIEKIRS 206
+ S +A+VHFYG KLQVQERMTRQIAE + + +VV EA H CMISRGIEK S
Sbjct: 368 VGSSLMKAIVHFYGFKLQVQERMTRQIAETLSPLVGGDVIVVAEAGHTCMISRGIEKFGS 427
Query: 207 NTATIAVLGQFLADPSAKACFLQNV 281
+TATIAVLG+F +D SA+A FL +
Sbjct: 428 STATIAVLGRFSSDNSARAMFLDKI 452
>sptr|Q90WT2|Q90WT2 GTP cyclohydrolase I.
Length = 240
Score = 71.6 bits (174), Expect = 5e-12
Identities = 37/80 (46%), Positives = 53/80 (66%), Gaps = 1/80 (1%)
Frame = +3
Query: 36 SHFQALVHFYGCKLQVQERMTRQIAEAVY-SVSHNGAMVVVEANHICMISRGIEKIRSNT 212
S +V Y +LQVQER+T+QIA A+ ++ G VV+EA H+CM+ RG++K+ S+T
Sbjct: 154 SKLARIVEIYSRRLQVQERLTKQIAMAISEALQPVGVAVVIEAAHMCMVMRGVQKMNSHT 213
Query: 213 ATIAVLGQFLADPSAKACFL 272
T A+LG F DP A+ FL
Sbjct: 214 VTSAMLGVFREDPKAREEFL 233
>sw|Q19980|GCH1_CAEEL Probable GTP cyclohydrolase I (EC 3.5.4.16)
(GTP-CH-I).
Length = 223
Score = 71.6 bits (174), Expect = 5e-12
Identities = 36/80 (45%), Positives = 53/80 (66%), Gaps = 1/80 (1%)
Frame = +3
Query: 36 SHFQALVHFYGCKLQVQERMTRQIAEA-VYSVSHNGAMVVVEANHICMISRGIEKIRSNT 212
S +V + +LQVQER+T+QIA A V +V +G VV+EA+H+CM+ RG++KI ++T
Sbjct: 138 SKLARIVEMFSRRLQVQERLTKQIATAMVQAVQPSGVAVVIEASHMCMVMRGVQKINAST 197
Query: 213 ATIAVLGQFLADPSAKACFL 272
T +LG F DP + FL
Sbjct: 198 TTSCMLGVFRDDPKTREEFL 217
>sw|O61573|GCH1_OSTOS GTP cyclohydrolase I (EC 3.5.4.16) (GTP-CH-I).
Length = 213
Score = 70.5 bits (171), Expect = 1e-11
Identities = 36/80 (45%), Positives = 52/80 (65%), Gaps = 1/80 (1%)
Frame = +3
Query: 36 SHFQALVHFYGCKLQVQERMTRQIAEA-VYSVSHNGAMVVVEANHICMISRGIEKIRSNT 212
S +V + +LQVQER+T+QIA A V +V G VV+EA+H+CM+ RG++KI + T
Sbjct: 129 SKLARIVEMFSRRLQVQERLTKQIATAMVQAVQPAGVAVVIEASHMCMVMRGVQKINATT 188
Query: 213 ATIAVLGQFLADPSAKACFL 272
+T +LG F DP + FL
Sbjct: 189 STSCMLGVFRDDPKTREEFL 208
>sw|P50141|GCH1_CHICK GTP cyclohydrolase I (EC 3.5.4.16) (GTP-CH-I).
Length = 236
Score = 69.7 bits (169), Expect = 2e-11
Identities = 36/80 (45%), Positives = 51/80 (63%), Gaps = 1/80 (1%)
Frame = +3
Query: 36 SHFQALVHFYGCKLQVQERMTRQIAEAVY-SVSHNGAMVVVEANHICMISRGIEKIRSNT 212
S +V Y +LQVQER+T+QIA A+ ++ G VV+EA H+CM+ RG++K+ S T
Sbjct: 152 SKLARIVEIYSRRLQVQERLTKQIAIAITEALQPAGVGVVIEATHMCMVMRGVQKMNSKT 211
Query: 213 ATIAVLGQFLADPSAKACFL 272
AT +LG F DP + FL
Sbjct: 212 ATSTMLGVFREDPKTREEFL 231
>sptrnew|AAH05643|AAH05643 GTP cyclohydrolase 1.
Length = 241
Score = 68.6 bits (166), Expect = 4e-11
Identities = 35/80 (43%), Positives = 50/80 (62%), Gaps = 1/80 (1%)
Frame = +3
Query: 36 SHFQALVHFYGCKLQVQERMTRQIAEAVY-SVSHNGAMVVVEANHICMISRGIEKIRSNT 212
S +V Y +LQVQER+T+QIA A+ ++ G VV+EA H+CM+ RG++K+ S T
Sbjct: 157 SKLARIVEIYSRRLQVQERLTKQIAVAITEALQPAGVGVVIEATHMCMVMRGVQKMNSKT 216
Query: 213 ATIAVLGQFLADPSAKACFL 272
T +LG F DP + FL
Sbjct: 217 VTSTMLGVFREDPKTREEFL 236
>sw|P22288|GCH1_RAT GTP cyclohydrolase I precursor (EC 3.5.4.16)
(GTP-CH-I).
Length = 241
Score = 68.6 bits (166), Expect = 4e-11
Identities = 35/80 (43%), Positives = 50/80 (62%), Gaps = 1/80 (1%)
Frame = +3
Query: 36 SHFQALVHFYGCKLQVQERMTRQIAEAVY-SVSHNGAMVVVEANHICMISRGIEKIRSNT 212
S +V Y +LQVQER+T+QIA A+ ++ G VV+EA H+CM+ RG++K+ S T
Sbjct: 157 SKLARIVEIYSRRLQVQERLTKQIAVAITEALQPAGVGVVIEATHMCMVMRGVQKMNSKT 216
Query: 213 ATIAVLGQFLADPSAKACFL 272
T +LG F DP + FL
Sbjct: 217 VTSTMLGVFREDPKTREEFL 236
>sw|Q05915|GCH1_MOUSE GTP cyclohydrolase I precursor (EC 3.5.4.16)
(GTP-CH-I).
Length = 241
Score = 68.6 bits (166), Expect = 4e-11
Identities = 35/80 (43%), Positives = 50/80 (62%), Gaps = 1/80 (1%)
Frame = +3
Query: 36 SHFQALVHFYGCKLQVQERMTRQIAEAVY-SVSHNGAMVVVEANHICMISRGIEKIRSNT 212
S +V Y +LQVQER+T+QIA A+ ++ G VV+EA H+CM+ RG++K+ S T
Sbjct: 157 SKLARIVEIYSRRLQVQERLTKQIAVAITEALQPAGVGVVIEATHMCMVMRGVQKMNSKT 216
Query: 213 ATIAVLGQFLADPSAKACFL 272
T +LG F DP + FL
Sbjct: 217 VTSTMLGVFREDPKTREEFL 236
>sptrnew|AAH25415|AAH25415 GTP cyclohydrolase 1 (Dopa-responsive
dystonia).
Length = 250
Score = 68.6 bits (166), Expect = 4e-11
Identities = 36/80 (45%), Positives = 50/80 (62%), Gaps = 1/80 (1%)
Frame = +3
Query: 36 SHFQALVHFYGCKLQVQERMTRQIAEAVY-SVSHNGAMVVVEANHICMISRGIEKIRSNT 212
S +V Y +LQVQER+T+QIA A+ ++ G VVVEA H+CM+ RG++K+ S T
Sbjct: 166 SKLARIVEIYSRRLQVQERLTKQIAVAITEALRPAGVGVVVEATHMCMVMRGVQKMNSKT 225
Query: 213 ATIAVLGQFLADPSAKACFL 272
T +LG F DP + FL
Sbjct: 226 VTSTMLGVFREDPKTREEFL 245
>sw|P30793|GCH1_HUMAN GTP cyclohydrolase I (EC 3.5.4.16) (GTP-CH-I).
Length = 250
Score = 68.6 bits (166), Expect = 4e-11
Identities = 36/80 (45%), Positives = 50/80 (62%), Gaps = 1/80 (1%)
Frame = +3
Query: 36 SHFQALVHFYGCKLQVQERMTRQIAEAVY-SVSHNGAMVVVEANHICMISRGIEKIRSNT 212
S +V Y +LQVQER+T+QIA A+ ++ G VVVEA H+CM+ RG++K+ S T
Sbjct: 166 SKLARIVEIYSRRLQVQERLTKQIAVAITEALRPAGVGVVVEATHMCMVMRGVQKMNSKT 225
Query: 213 ATIAVLGQFLADPSAKACFL 272
T +LG F DP + FL
Sbjct: 226 VTSTMLGVFREDPKTREEFL 245
>sw|P48596|GCH1_DROME GTP cyclohydrolase I (EC 3.5.4.16) (GTP-CH-I)
(Punch protein).
Length = 324
Score = 68.6 bits (166), Expect = 4e-11
Identities = 38/83 (45%), Positives = 50/83 (60%), Gaps = 1/83 (1%)
Frame = +3
Query: 36 SHFQALVHFYGCKLQVQERMTRQIAEAV-YSVSHNGAMVVVEANHICMISRGIEKIRSNT 212
S +V + +LQVQER+T+QIA AV +V G VVVE H+CM+ RG++KI S T
Sbjct: 239 SKLARIVEIFSRRLQVQERLTKQIAVAVTQAVQPAGVAVVVEGVHMCMVMRGVQKINSKT 298
Query: 213 ATIAVLGQFLADPSAKACFLQNV 281
T +LG F DP + FL V
Sbjct: 299 VTSTMLGVFRDDPKTREEFLNLV 321
>sptr|Q8IZH9|Q8IZH9 GTP cyclohydrolase I type IV (EC 3.5.4.16)
(Fragment).
Length = 177
Score = 68.6 bits (166), Expect = 4e-11
Identities = 36/80 (45%), Positives = 50/80 (62%), Gaps = 1/80 (1%)
Frame = +3
Query: 36 SHFQALVHFYGCKLQVQERMTRQIAEAVY-SVSHNGAMVVVEANHICMISRGIEKIRSNT 212
S +V Y +LQVQER+T+QIA A+ ++ G VVVEA H+CM+ RG++K+ S T
Sbjct: 93 SKLARIVEIYSRRLQVQERLTKQIAVAITEALRPAGVGVVVEATHMCMVMRGVQKMNSKT 152
Query: 213 ATIAVLGQFLADPSAKACFL 272
T +LG F DP + FL
Sbjct: 153 VTSTMLGVFREDPKTREEFL 172
>sw|Q8DJB8|GCH1_SYNEL GTP cyclohydrolase I (EC 3.5.4.16) (GTP-CH-I).
Length = 229
Score = 67.4 bits (163), Expect = 1e-10
Identities = 37/83 (44%), Positives = 51/83 (61%), Gaps = 1/83 (1%)
Frame = +3
Query: 27 IDRSHFQALVHFYGCKLQVQERMTRQIAEAVYSV-SHNGAMVVVEANHICMISRGIEKIR 203
I S +V Y +LQVQER+TRQ+AEA+ +V G VV+EA H+CM+ RG++K
Sbjct: 138 IGLSKLARVVEMYARRLQVQERLTRQVAEAIETVLDPKGVAVVMEATHMCMVMRGVQKPG 197
Query: 204 SNTATIAVLGQFLADPSAKACFL 272
S T T ++LG F D + FL
Sbjct: 198 SWTVTSSMLGVFREDQKTREEFL 220
>sptrnew|EAA11896|EAA11896 AgCP6428 (Fragment).
Length = 344
Score = 67.4 bits (163), Expect = 1e-10
Identities = 35/80 (43%), Positives = 49/80 (61%), Gaps = 1/80 (1%)
Frame = +3
Query: 36 SHFQALVHFYGCKLQVQERMTRQIAEAV-YSVSHNGAMVVVEANHICMISRGIEKIRSNT 212
S +V + +LQVQER+T+QIA AV +V G V++E H+CM+ RG++KI S T
Sbjct: 259 SKLARIVEIFSRRLQVQERLTKQIAVAVTQAVQPAGVAVIIEGVHMCMVMRGVQKINSKT 318
Query: 213 ATIAVLGQFLADPSAKACFL 272
T +LG F DP + FL
Sbjct: 319 VTSTMLGVFRDDPKTREEFL 338
>sw|Q8YLL1|GC11_ANASP GTP cyclohydrolase I 1 (EC 3.5.4.16) (GTP-CH-I
1).
Length = 235
Score = 66.6 bits (161), Expect = 2e-10
Identities = 36/79 (45%), Positives = 50/79 (63%), Gaps = 1/79 (1%)
Frame = +3
Query: 36 SHFQALVHFYGCKLQVQERMTRQIAEAVYSVSH-NGAMVVVEANHICMISRGIEKIRSNT 212
S +V Y +LQVQER+TRQIAEAV ++ G VV+EA+H+CM+ RG++K S T
Sbjct: 146 SKLARIVEMYSRRLQVQERLTRQIAEAVQTILEPQGVAVVMEASHMCMVMRGVQKPGSWT 205
Query: 213 ATIAVLGQFLADPSAKACF 269
T A+LG F + + F
Sbjct: 206 VTSAMLGVFQEEQKTREEF 224
>sw|Q9RYB4|GCH1_DEIRA GTP cyclohydrolase I (EC 3.5.4.16) (GTP-CH-I).
Length = 216
Score = 66.6 bits (161), Expect = 2e-10
Identities = 35/83 (42%), Positives = 52/83 (62%), Gaps = 1/83 (1%)
Frame = +3
Query: 36 SHFQALVHFYGCKLQVQERMTRQIAEAVYSV-SHNGAMVVVEANHICMISRGIEKIRSNT 212
S F +V Y +LQVQER+T Q+A+AV + + G V++E H+CM RG++K S+T
Sbjct: 127 SKFARIVDLYSRRLQVQERITTQVADAVEELLAPKGVAVLMEGIHLCMAMRGVQKQNSST 186
Query: 213 ATIAVLGQFLADPSAKACFLQNV 281
T A+ G F +DP +A F+ V
Sbjct: 187 TTSAMRGLFRSDPRTRAEFMSAV 209
>sw|Q8KEA8|GCH1_CHLTE GTP cyclohydrolase I (EC 3.5.4.16) (GTP-CH-I).
Length = 224
Score = 66.6 bits (161), Expect = 2e-10
Identities = 34/87 (39%), Positives = 55/87 (63%), Gaps = 1/87 (1%)
Frame = +3
Query: 18 GEGIDRSHFQALVHFYGCKLQVQERMTRQIAEAVYSVSH-NGAMVVVEANHICMISRGIE 194
G+ + S +V + +LQVQER+T+QI +A+ +V + G VV+EA H+CM+ RG+E
Sbjct: 133 GKIVGLSKIPRVVEVFARRLQVQERLTQQIRDAIQNVLNPRGVAVVIEATHMCMVMRGVE 192
Query: 195 KIRSNTATIAVLGQFLADPSAKACFLQ 275
K + T T A+ G F+ S ++ FL+
Sbjct: 193 KQNAVTTTSAMSGDFMTSQSTRSEFLR 219
>sw|Q54769|GCH1_SYNP7 GTP cyclohydrolase I (EC 3.5.4.16) (GTP-CH-I).
Length = 213
Score = 66.2 bits (160), Expect = 2e-10
Identities = 36/80 (45%), Positives = 49/80 (61%), Gaps = 1/80 (1%)
Frame = +3
Query: 36 SHFQALVHFYGCKLQVQERMTRQIAEAVYSV-SHNGAMVVVEANHICMISRGIEKIRSNT 212
S +V Y +LQVQER+TRQIAE+V + G VV+EA H+CM+ RG++K S T
Sbjct: 123 SKLARIVEMYSRRLQVQERLTRQIAESVQEILDPQGVAVVMEATHMCMVMRGVQKPGSWT 182
Query: 213 ATIAVLGQFLADPSAKACFL 272
T A++G F D + FL
Sbjct: 183 VTSAMVGVFQEDQRTREEFL 202
>sw|Q9I351|GC12_PSEAE GTP cyclohydrolase I 2 (EC 3.5.4.16) (GTP-CH-I
2).
Length = 181
Score = 66.2 bits (160), Expect = 2e-10
Identities = 35/87 (40%), Positives = 55/87 (63%), Gaps = 1/87 (1%)
Frame = +3
Query: 15 SGEGIDRSHFQALVHFYGCKLQVQERMTRQIAEAVYSVSHN-GAMVVVEANHICMISRGI 191
+G+ + S +V Y +LQ+QE M+RQIAEAV V+ G VV++A H+CM+ RG+
Sbjct: 91 NGKVLGLSKVARIVDMYARRLQIQENMSRQIAEAVQQVTGALGVAVVIQAQHMCMMMRGV 150
Query: 192 EKIRSNTATIAVLGQFLADPSAKACFL 272
EK S+ T +LG+F + + ++ FL
Sbjct: 151 EKQNSSMVTSVMLGEFRDNAATRSEFL 177
>sptr|Q83DE3|Q83DE3 GTP cyclohydrolase I.
Length = 184
Score = 66.2 bits (160), Expect = 2e-10
Identities = 35/87 (40%), Positives = 52/87 (59%), Gaps = 1/87 (1%)
Frame = +3
Query: 15 SGEGIDRSHFQALVHFYGCKLQVQERMTRQIAEAVYSVSH-NGAMVVVEANHICMISRGI 191
SG+ I S +V + +LQVQE +T+QIAEA+ + + G V++EA H+CM+ RG+
Sbjct: 93 SGKIIGISKLARIVDMFAKRLQVQENLTKQIAEAILTATEAKGVGVIIEAKHLCMMMRGV 152
Query: 192 EKIRSNTATIAVLGQFLADPSAKACFL 272
EK S T +LG F D ++ FL
Sbjct: 153 EKQNSEMTTSVMLGTFRKDDRTRSEFL 179
>sptr|Q96T74|Q96T74 GTP cyclohydrolase I (Fragment).
Length = 78
Score = 65.9 bits (159), Expect = 3e-10
Identities = 34/71 (47%), Positives = 47/71 (66%), Gaps = 1/71 (1%)
Frame = +3
Query: 63 YGCKLQVQERMTRQIAEAVY-SVSHNGAMVVVEANHICMISRGIEKIRSNTATIAVLGQF 239
Y +LQVQER+T+QIA A+ ++ G VVVEA H+CM+ RG++K+ S T T +LG F
Sbjct: 3 YSRRLQVQERLTKQIAVAITEALRPAGVGVVVEATHMCMVMRGVQKMNSKTVTSTMLGVF 62
Query: 240 LADPSAKACFL 272
DP + FL
Sbjct: 63 REDPKTREEFL 73
>sptr|Q884Q3|Q884Q3 GTP cyclohydrolase I.
Length = 181
Score = 65.5 bits (158), Expect = 4e-10
Identities = 34/87 (39%), Positives = 54/87 (62%), Gaps = 1/87 (1%)
Frame = +3
Query: 15 SGEGIDRSHFQALVHFYGCKLQVQERMTRQIAEAVYSVSH-NGAMVVVEANHICMISRGI 191
+G+ + S +V Y +LQ+QE ++RQIAEA+ V+ G VV+EA H+CM+ RG+
Sbjct: 91 NGKVLGLSKVARIVDMYARRLQIQENLSRQIAEAIQQVTGAQGVAVVIEAKHMCMMMRGV 150
Query: 192 EKIRSNTATIAVLGQFLADPSAKACFL 272
EK S T +LG+F + + ++ FL
Sbjct: 151 EKQNSAMITSVMLGEFRENAATRSEFL 177
>sptr|Q88LV4|Q88LV4 GTP cyclohydrolase I.
Length = 190
Score = 65.5 bits (158), Expect = 4e-10
Identities = 36/86 (41%), Positives = 53/86 (61%), Gaps = 1/86 (1%)
Frame = +3
Query: 18 GEGIDRSHFQALVHFYGCKLQVQERMTRQIAEAVYSVSHN-GAMVVVEANHICMISRGIE 194
G+ + S +V Y +LQ+QE ++RQIAEAV V+ G VVVEA H+CM+ RG+E
Sbjct: 101 GKVLGLSKVARIVDMYARRLQIQENLSRQIAEAVQQVTGAAGVAVVVEAKHMCMMMRGVE 160
Query: 195 KIRSNTATIAVLGQFLADPSAKACFL 272
K S T +LG+F + + ++ FL
Sbjct: 161 KQNSTMITSVMLGEFRDNAATRSEFL 186
>sw|Q55759|GCH1_SYNY3 GTP cyclohydrolase I (EC 3.5.4.16) (GTP-CH-I).
Length = 234
Score = 65.5 bits (158), Expect = 4e-10
Identities = 35/80 (43%), Positives = 49/80 (61%), Gaps = 1/80 (1%)
Frame = +3
Query: 36 SHFQALVHFYGCKLQVQERMTRQIAEAVYSV-SHNGAMVVVEANHICMISRGIEKIRSNT 212
S +V + +LQVQER+TRQIAEAV + G VV+EA H+CM+ RG++K S T
Sbjct: 145 SKLARIVEMFSRRLQVQERLTRQIAEAVQEILDPQGVAVVMEATHMCMVMRGVQKPGSWT 204
Query: 213 ATIAVLGQFLADPSAKACFL 272
T A++G F + + FL
Sbjct: 205 VTSAMIGSFQNEQKTREEFL 224
>sw|Q9HYG8|GC11_PSEAE GTP cyclohydrolase I 1 (EC 3.5.4.16) (GTP-CH-I
1).
Length = 186
Score = 65.1 bits (157), Expect = 5e-10
Identities = 37/88 (42%), Positives = 52/88 (59%), Gaps = 1/88 (1%)
Frame = +3
Query: 15 SGEGIDRSHFQALVHFYGCKLQVQERMTRQIAEAVYSV-SHNGAMVVVEANHICMISRGI 191
+G+ + S +V + +LQ+QE +TRQIAEAV V S G VV+EA H+CM+ RG+
Sbjct: 93 TGKVLGLSKVARIVDMFARRLQIQENLTRQIAEAVRQVTSAAGVAVVIEAQHMCMMMRGV 152
Query: 192 EKIRSNTATIAVLGQFLADPSAKACFLQ 275
EK S T +LG F + + FLQ
Sbjct: 153 EKQNSQMFTSVMLGAFRDSNTTRQEFLQ 180
>sw|O69531|GCH1_MYCLE GTP cyclohydrolase I (EC 3.5.4.16) (GTP-CH-I).
Length = 205
Score = 63.9 bits (154), Expect = 1e-09
Identities = 35/77 (45%), Positives = 48/77 (62%), Gaps = 1/77 (1%)
Frame = +3
Query: 36 SHFQALVHFYGCKLQVQERMTRQIAEAVYS-VSHNGAMVVVEANHICMISRGIEKIRSNT 212
S LV Y + QVQER+T QIA+A+ S + G ++VVEA H+CM RG+ K + T
Sbjct: 120 SKIARLVDLYAKRPQVQERLTSQIADALVSKLDPRGVIIVVEAEHLCMAMRGVRKPGAIT 179
Query: 213 ATIAVLGQFLADPSAKA 263
T AV GQF D +++A
Sbjct: 180 TTSAVRGQFKTDAASRA 196
>sptr|Q8GLS8|Q8GLS8 GTP cyclohydrolase I (EC 3.5.4.16).
Length = 227
Score = 63.9 bits (154), Expect = 1e-09
Identities = 37/84 (44%), Positives = 51/84 (60%), Gaps = 1/84 (1%)
Frame = +3
Query: 36 SHFQALVHFYGCKLQVQERMTRQIAEAVY-SVSHNGAMVVVEANHICMISRGIEKIRSNT 212
S LV Y + QVQER+T Q+A+AV + GA+VVVEA H+CM RGI K ++T
Sbjct: 142 SKLARLVDLYAKRPQVQERLTSQVADAVMRKLDPRGAIVVVEAEHLCMAMRGIRKPGAST 201
Query: 213 ATIAVLGQFLADPSAKACFLQNVL 284
T AV G + PS+++ L +L
Sbjct: 202 TTSAVRGLLQSSPSSRSEALDLIL 225
Database: /db/trembl-ebi/tmp/swall
Posted date: Sep 26, 2003 8:29 PM
Number of letters in database: 406,886,216
Number of sequences in database: 1,272,877
Lambda K H
0.318 0.135 0.401
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 429,691,707
Number of Sequences: 1272877
Number of extensions: 9210004
Number of successful extensions: 22481
Number of sequences better than 10.0: 30
Number of HSP's better than 10.0 without gapping: 21586
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 22336
length of database: 406,886,216
effective HSP length: 115
effective length of database: 260,505,361
effective search space used: 17193353826
frameshift window, decay const: 50, 0.1
T: 12
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)

