BLASTX 2.2.6 [Apr-09-2003]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= 3204272.2.1
(739 letters)
Database: /db/trembl-ebi/tmp/swall
1,272,877 sequences; 406,886,216 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
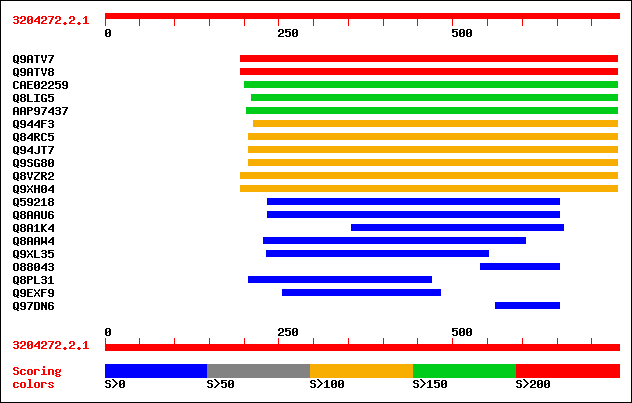
sptr|Q9ATV7|Q9ATV7 Arabinoxylan arabinofuranohydrolase isoenzyme... 252 3e-66
sptr|Q9ATV8|Q9ATV8 Arabinoxylan arabinofuranohydrolase isoenzyme... 228 6e-59
sptrnew|CAE02259|CAE02259 OSJNBb0058J09.6 protein. 186 3e-46
sptr|Q8LIG5|Q8LIG5 Putative arabinoxylan narabinofuranohydrolase... 164 1e-39
sptrnew|AAP97437|AAP97437 Alpha-L-arabinofuranosidase. 158 6e-38
sptr|Q944F3|Q944F3 Arabinosidase ARA-1. 145 4e-34
sptr|Q84RC5|Q84RC5 Alpha-L-arabinofuranosidase. 140 1e-32
sptr|Q94JT7|Q94JT7 AT3g10740/T7M13_18. 140 1e-32
sptr|Q9SG80|Q9SG80 Putative alpha-L-arabinofuranosidase. 140 1e-32
sptr|Q8VZR2|Q8VZR2 Putative arabinosidase (Alpha-L-arabinofurano... 125 6e-28
sptr|Q9XH04|Q9XH04 T1N24.13 protein. 125 6e-28
sptr|Q59218|Q59218 Arabinosidase (EC 3.2.1.55). 46 6e-04
sptr|Q8AAU6|Q8AAU6 Alpha-L-arabinofuranosidase A precursor. 45 0.001
sptr|Q8A1K4|Q8A1K4 Alpha-L-arabinofuranosidase A precursor. 38 0.16
sptr|Q8AAW4|Q8AAW4 Alpha-L-arabinofuranosidase. 35 1.3
sptr|Q9XL35|Q9XL35 NADH dehydrogenase subunit V. 35 1.3
sptr|O88043|O88043 Putative secreted arabinosidase. 34 2.3
sptr|Q8PL31|Q8PL31 Flagellar protein. 33 5.0
sptr|Q9EXF9|Q9EXF9 Proline-rich protein (Lmo0320 protein). 32 6.6
sptr|Q97DN6|Q97DN6 Probable alpha-arabinofuranosidase. 32 6.6
>sptr|Q9ATV7|Q9ATV7 Arabinoxylan arabinofuranohydrolase isoenzyme
AXAH-II.
Length = 656
Score = 252 bits (644), Expect = 3e-66
Identities = 135/200 (67%), Positives = 142/200 (71%), Gaps = 19/200 (9%)
Frame = -3
Query: 737 AVWRGDAGRGXXXXXXXXXXXLTGLEKNR-------------------WNPDAIVFNSWQ 615
AVWR DAGRG LTGLEKN WNPDAIVFNSWQ
Sbjct: 457 AVWRTDAGRGSLLGSLAEAAFLTGLEKNSDIVHMASYAPLFVNDNDQTWNPDAIVFNSWQ 516
Query: 614 HYGTPSYWMQTLFRESSGAMFHPTTIXXXXXXXXXXXAITWQDSENSFLRVKVINFGSDA 435
YGTPSYWMQ FRESSGAM HP TI AITWQDS N+FLRVK++NFGSD
Sbjct: 517 QYGTPSYWMQKFFRESSGAMIHPITISSSYSGSLAASAITWQDSGNNFLRVKIVNFGSDT 576
Query: 434 VTLTISTSGLQASINALGSTATVLTSADVMDENSFSNPTKVVPVKSDLSNAAEQMQVTLA 255
V+LTIS SGLQASINALGS ATVLTS++V DENSFSNPTKVVPV S L NAAEQMQVTLA
Sbjct: 577 VSLTISVSGLQASINALGSNATVLTSSNVKDENSFSNPTKVVPVTSQLHNAAEQMQVTLA 636
Query: 254 PRSFSSFDLALAQSKLVAEM 195
SFSSFDLALAQS+LVAEM
Sbjct: 637 AHSFSSFDLALAQSELVAEM 656
>sptr|Q9ATV8|Q9ATV8 Arabinoxylan arabinofuranohydrolase isoenzyme
AXAH-I.
Length = 658
Score = 228 bits (581), Expect = 6e-59
Identities = 121/200 (60%), Positives = 135/200 (67%), Gaps = 19/200 (9%)
Frame = -3
Query: 737 AVWRGDAGRGXXXXXXXXXXXLTGLEKNR-------------------WNPDAIVFNSWQ 615
AV DAGRG LTGLEKN WNPDAIVFNSWQ
Sbjct: 459 AVTGNDAGRGSLLASLAEAAFLTGLEKNSDVVQMASYAPLFINDNDRTWNPDAIVFNSWQ 518
Query: 614 HYGTPSYWMQTLFRESSGAMFHPTTIXXXXXXXXXXXAITWQDSENSFLRVKVINFGSDA 435
YGTPSYWMQTLFRESSGA HP ++ AITW D ENSFLRVK++NFG DA
Sbjct: 519 QYGTPSYWMQTLFRESSGATVHPISLSSSCSGLLAASAITWHDDENSFLRVKIVNFGPDA 578
Query: 434 VTLTISTSGLQASINALGSTATVLTSADVMDENSFSNPTKVVPVKSDLSNAAEQMQVTLA 255
V LTIS +GLQ SINA GSTATVLTS VMDENSF+NP KVVPV +L +AAE+MQVTL
Sbjct: 579 VGLTISATGLQGSINAFGSTATVLTSGGVMDENSFANPNKVVPVTVELRDAAEEMQVTLP 638
Query: 254 PRSFSSFDLALAQSKLVAEM 195
P S ++FDLALAQS+LVA++
Sbjct: 639 PHSLTAFDLALAQSRLVADV 658
>sptrnew|CAE02259|CAE02259 OSJNBb0058J09.6 protein.
Length = 790
Score = 186 bits (472), Expect = 3e-46
Identities = 99/198 (50%), Positives = 120/198 (60%), Gaps = 19/198 (9%)
Frame = -3
Query: 737 AVWRGDAGRGXXXXXXXXXXXLTGLEKNR-------------------WNPDAIVFNSWQ 615
AV GRG LTGLEKN WNP A+VFNSW+
Sbjct: 593 AVSSNGVGRGTLLASLAEAAFLTGLEKNSDVVQMASYAPLFMNDNDLSWNPAAVVFNSWK 652
Query: 614 HYGTPSYWMQTLFRESSGAMFHPTTIXXXXXXXXXXXAITWQDSENSFLRVKVINFGSDA 435
YG+PSYWMQT+FRESSGA+ HP TI AITW+ S +SFLRVK++N GS+
Sbjct: 653 QYGSPSYWMQTIFRESSGAVLHPVTINSMYSNSLAASAITWKASNSSFLRVKIVNIGSNP 712
Query: 434 VTLTISTSGLQASINALGSTATVLTSADVMDENSFSNPTKVVPVKSDLSNAAEQMQVTLA 255
V L +ST+GL+A +N ST T+LTS ++ DENSFS PT VVPV +L NA E+M L
Sbjct: 713 VNLIVSTTGLEALVNMRKSTITILTSKNLSDENSFSKPTNVVPVTRELPNAGEEMFAFLG 772
Query: 254 PRSFSSFDLALAQSKLVA 201
P SF+SFDLAL Q K V+
Sbjct: 773 PYSFTSFDLALGQQKHVS 790
>sptr|Q8LIG5|Q8LIG5 Putative arabinoxylan narabinofuranohydrolase
isoenzyme AXAH-I.
Length = 664
Score = 164 bits (414), Expect = 1e-39
Identities = 90/197 (45%), Positives = 115/197 (58%), Gaps = 21/197 (10%)
Frame = -3
Query: 737 AVWRGDAGRGXXXXXXXXXXXLTGLEKN-------------------RWNPDAIVFNSWQ 615
AV DAG+G L GLE+N RW+PDAIVFNSWQ
Sbjct: 464 AVTGKDAGKGTLVEALAEAAFLVGLERNSDVVEMASCAPLFVNDNDRRWSPDAIVFNSWQ 523
Query: 614 HYGTPSYWMQTLFRESSGAMFHPTTIXXXXXXXXXXXAITWQDSEN--SFLRVKVINFGS 441
+YG P+YWM F++S GA FHP+ I AITWQ+S++ ++L++K++NFG+
Sbjct: 524 NYGCPNYWMLHFFKDSCGATFHPSNIQISSYNQLVASAITWQNSKDKSTYLKIKLVNFGN 583
Query: 440 DAVTLTISTSGLQASINALGSTATVLTSADVMDENSFSNPTKVVPVKSDLSNAAEQMQVT 261
AV L+IS SGL I + GS TVLTS+ +DENSF P KV PV S + NA EQM V
Sbjct: 584 QAVNLSISVSGLDEGIKSSGSKKTVLTSSGPLDENSFQQPQKVAPVSSPVDNANEQMGVL 643
Query: 260 LAPRSFSSFDLALAQSK 210
+ P S +SFDL L SK
Sbjct: 644 VDPYSLTSFDLLLQPSK 660
>sptrnew|AAP97437|AAP97437 Alpha-L-arabinofuranosidase.
Length = 675
Score = 158 bits (400), Expect = 6e-38
Identities = 90/200 (45%), Positives = 119/200 (59%), Gaps = 22/200 (11%)
Frame = -3
Query: 737 AVWRGDAGRGXXXXXXXXXXXLTGLEKN-------------------RWNPDAIVFNSWQ 615
AV DAGRG L GLEKN RWNPDAIVF+S Q
Sbjct: 463 AVTGKDAGRGSLLAALAEAGFLIGLEKNSDIVEMASYAPLFVNTHDRRWNPDAIVFDSSQ 522
Query: 614 HYGTPSYWMQTLFRESSGAMFHPTTIXXXXXXXXXXXAITWQDS--ENSFLRVKVINFGS 441
YGTPSYW+Q LF ESSGA + +T+ AI+WQ+S EN++LR+KV+N G+
Sbjct: 523 LYGTPSYWVQCLFNESSGATLYNSTLQMNSSTSLLASAISWQNSENENTYLRIKVVNLGT 582
Query: 440 DAVTLTISTSGLQA-SINALGSTATVLTSADVMDENSFSNPTKVVPVKSDLSNAAEQMQV 264
V L + GL+ S++ GST TVLTS + MDENSF+ P KV+P +S L +A E+M+V
Sbjct: 583 YTVNLKVFVDGLEPNSVSLSGSTKTVLTSNNQMDENSFNEPKKVIPKRSLLESAGEEMEV 642
Query: 263 TLAPRSFSSFDLALAQSKLV 204
++PRSF+S DL + S ++
Sbjct: 643 VISPRSFTSIDLLMESSDVI 662
>sptr|Q944F3|Q944F3 Arabinosidase ARA-1.
Length = 674
Score = 145 bits (367), Expect = 4e-34
Identities = 87/198 (43%), Positives = 111/198 (56%), Gaps = 23/198 (11%)
Frame = -3
Query: 737 AVWRGDAGRGXXXXXXXXXXXLTGLEKN-------------------RWNPDAIVFNSWQ 615
AV DAG+G L G+EKN RWNPDAIVF S Q
Sbjct: 462 AVTGNDAGKGSLLAALGEAGFLIGVEKNSEAIEMASYAPLFVNDNDRRWNPDAIVFTSSQ 521
Query: 614 HYGTPSYWMQTLFRESSGAMFHPTTIXXXXXXXXXXXAITWQDS--ENSFLRVKVINFGS 441
YGTPSYWMQ F+ES+GA +++ AITW++S N +LR+KV+NFG+
Sbjct: 522 MYGTPSYWMQHFFKESNGATLLSSSLQANPSNSLIASAITWRNSLDNNDYLRIKVVNFGT 581
Query: 440 DAVTLTISTSGL-QASINAL-GSTATVLTSADVMDENSFSNPTKVVPVKSDLSNAAEQMQ 267
AV IS +GL Q S+ L G+ T LTS +VMDENSF P KV+PVK+ + ++ M
Sbjct: 582 TAVITKISLTGLGQNSLETLFGAVMTELTSNNVMDENSFREPNKVIPVKTQVEKVSDNMD 641
Query: 266 VTLAPRSFSSFDLALAQS 213
V LAPRS +S D L +S
Sbjct: 642 VVLAPRSLNSIDFLLRKS 659
>sptr|Q84RC5|Q84RC5 Alpha-L-arabinofuranosidase.
Length = 678
Score = 140 bits (354), Expect = 1e-32
Identities = 83/197 (42%), Positives = 107/197 (54%), Gaps = 20/197 (10%)
Frame = -3
Query: 737 AVWRGDAGRGXXXXXXXXXXXLTGLEKN-------------------RWNPDAIVFNSWQ 615
AV DAG G L GLEKN RWNPDAIVFNS
Sbjct: 467 AVTGKDAGTGSLLASLAEAAFLIGLEKNSDIVDMASYAPLFVNTNDRRWNPDAIVFNSSH 526
Query: 614 HYGTPSYWMQTLFRESSGAMFHPTTIXXXXXXXXXXXAITWQDSENSFLRVKVINFGSDA 435
YGTPSYW+Q F ESSGA +T+ I+W+++ ++R+K +NFG+++
Sbjct: 527 LYGTPSYWVQRFFAESSGATLLTSTLKGNSTSLVASA-ISWKNNGKDYIRIKAVNFGANS 585
Query: 434 VTLTISTSGLQASINAL-GSTATVLTSADVMDENSFSNPTKVVPVKSDLSNAAEQMQVTL 258
+ + +GL ++ + GS TVLTS +VMDENSFS P KVVP +S L A E M V L
Sbjct: 586 ENMQVLVTGLDPNVMRVSGSKKTVLTSTNVMDENSFSQPEKVVPHESLLELAEEDMTVVL 645
Query: 257 APRSFSSFDLALAQSKL 207
P SFSSFDL +K+
Sbjct: 646 PPHSFSSFDLLKESAKI 662
>sptr|Q94JT7|Q94JT7 AT3g10740/T7M13_18.
Length = 678
Score = 140 bits (354), Expect = 1e-32
Identities = 83/197 (42%), Positives = 107/197 (54%), Gaps = 20/197 (10%)
Frame = -3
Query: 737 AVWRGDAGRGXXXXXXXXXXXLTGLEKN-------------------RWNPDAIVFNSWQ 615
AV DAG G L GLEKN RWNPDAIVFNS
Sbjct: 467 AVTGKDAGTGSLLASLAEAAFLIGLEKNSDIVEMASYAPLFVNTNDRRWNPDAIVFNSSH 526
Query: 614 HYGTPSYWMQTLFRESSGAMFHPTTIXXXXXXXXXXXAITWQDSENSFLRVKVINFGSDA 435
YGTPSYW+Q F ESSGA +T+ I+W+++ ++R+K +NFG+++
Sbjct: 527 LYGTPSYWVQRFFAESSGATLLTSTLKGNSTSLVASA-ISWKNNGKDYIRIKAVNFGANS 585
Query: 434 VTLTISTSGLQASINAL-GSTATVLTSADVMDENSFSNPTKVVPVKSDLSNAAEQMQVTL 258
+ + +GL ++ + GS TVLTS +VMDENSFS P KVVP +S L A E M V L
Sbjct: 586 ENMQVLVTGLDPNVMRVSGSKKTVLTSTNVMDENSFSQPEKVVPHESLLELAEEDMTVVL 645
Query: 257 APRSFSSFDLALAQSKL 207
P SFSSFDL +K+
Sbjct: 646 PPHSFSSFDLLKESAKI 662
>sptr|Q9SG80|Q9SG80 Putative alpha-L-arabinofuranosidase.
Length = 678
Score = 140 bits (354), Expect = 1e-32
Identities = 83/197 (42%), Positives = 107/197 (54%), Gaps = 20/197 (10%)
Frame = -3
Query: 737 AVWRGDAGRGXXXXXXXXXXXLTGLEKN-------------------RWNPDAIVFNSWQ 615
AV DAG G L GLEKN RWNPDAIVFNS
Sbjct: 467 AVTGKDAGTGSLLASLAEAAFLIGLEKNSDIVEMASYAPLFVNTNDRRWNPDAIVFNSSH 526
Query: 614 HYGTPSYWMQTLFRESSGAMFHPTTIXXXXXXXXXXXAITWQDSENSFLRVKVINFGSDA 435
YGTPSYW+Q F ESSGA +T+ I+W+++ ++R+K +NFG+++
Sbjct: 527 LYGTPSYWVQRFFAESSGATLLTSTLKGNSTSLVASA-ISWKNNGKDYIRIKAVNFGANS 585
Query: 434 VTLTISTSGLQASINAL-GSTATVLTSADVMDENSFSNPTKVVPVKSDLSNAAEQMQVTL 258
+ + +GL ++ + GS TVLTS +VMDENSFS P KVVP +S L A E M V L
Sbjct: 586 ENMQVLVTGLDPNVMRVSGSKKTVLTSTNVMDENSFSQPEKVVPHESLLELAEEDMTVVL 645
Query: 257 APRSFSSFDLALAQSKL 207
P SFSSFDL +K+
Sbjct: 646 PPHSFSSFDLLKESAKI 662
>sptr|Q8VZR2|Q8VZR2 Putative arabinosidase
(Alpha-L-arabinofuranosidase) (Hypothetical protein
At5g26120).
Length = 674
Score = 125 bits (314), Expect = 6e-28
Identities = 83/201 (41%), Positives = 105/201 (52%), Gaps = 20/201 (9%)
Frame = -3
Query: 737 AVWRGDAGRGXXXXXXXXXXXLTGLEKN-------------------RWNPDAIVFNSWQ 615
AV + DA G L GLEKN RW PDAIVFNS
Sbjct: 466 AVNKADAKNGNLLAALGEAAFLLGLEKNSDIVEMVSYAPLFVNTNDRRWIPDAIVFNSSH 525
Query: 614 HYGTPSYWMQTLFRESSGAMFHPTTIXXXXXXXXXXXAITWQDSENSFLRVKVINFGSDA 435
YGTPSYW+Q F ESSGA +T+ AI++Q + ++++K +NFG +
Sbjct: 526 LYGTPSYWVQHFFTESSGATLLNSTL-KGKTSSVEASAISFQTNGKDYIQIKAVNFGEQS 584
Query: 434 VTLTISTSGLQASINALGSTATVLTSADVMDENSFSNPTKVVPVKSDLS-NAAEQMQVTL 258
V L ++ +GL A GS VLTSA VMDENSFSNP +VP +S L E + L
Sbjct: 585 VNLKVAVTGLMAKF--YGSKKKVLTSASVMDENSFSNPNMIVPQESLLEMTEQEDLMFVL 642
Query: 257 APRSFSSFDLALAQSKLVAEM 195
P SFSSFDL L +S+ V +M
Sbjct: 643 PPHSFSSFDL-LTESENVIKM 662
>sptr|Q9XH04|Q9XH04 T1N24.13 protein.
Length = 521
Score = 125 bits (314), Expect = 6e-28
Identities = 83/201 (41%), Positives = 105/201 (52%), Gaps = 20/201 (9%)
Frame = -3
Query: 737 AVWRGDAGRGXXXXXXXXXXXLTGLEKN-------------------RWNPDAIVFNSWQ 615
AV + DA G L GLEKN RW PDAIVFNS
Sbjct: 313 AVNKADAKNGNLLAALGEAAFLLGLEKNSDIVEMVSYAPLFVNTNDRRWIPDAIVFNSSH 372
Query: 614 HYGTPSYWMQTLFRESSGAMFHPTTIXXXXXXXXXXXAITWQDSENSFLRVKVINFGSDA 435
YGTPSYW+Q F ESSGA +T+ AI++Q + ++++K +NFG +
Sbjct: 373 LYGTPSYWVQHFFTESSGATLLNSTL-KGKTSSVEASAISFQTNGKDYIQIKAVNFGEQS 431
Query: 434 VTLTISTSGLQASINALGSTATVLTSADVMDENSFSNPTKVVPVKSDLS-NAAEQMQVTL 258
V L ++ +GL A GS VLTSA VMDENSFSNP +VP +S L E + L
Sbjct: 432 VNLKVAVTGLMAKF--YGSKKKVLTSASVMDENSFSNPNMIVPQESLLEMTEQEDLMFVL 489
Query: 257 APRSFSSFDLALAQSKLVAEM 195
P SFSSFDL L +S+ V +M
Sbjct: 490 PPHSFSSFDL-LTESENVIKM 509
>sptr|Q59218|Q59218 Arabinosidase (EC 3.2.1.55).
Length = 660
Score = 45.8 bits (107), Expect = 6e-04
Identities = 31/145 (21%), Positives = 60/145 (41%), Gaps = 5/145 (3%)
Frame = -3
Query: 653 RWNPDAIVFNSWQHYGTPSYWMQTLFRESSGAMFHPTT-----IXXXXXXXXXXXAITWQ 489
+W PD I F++ T SY++Q L+ ++ G P T + + +
Sbjct: 512 QWRPDMIWFDNLNSVRTTSYYVQQLYAQNKGTNVLPLTMNKKNVTGAEGQNGLFASAVYD 571
Query: 488 DSENSFLRVKVINFGSDAVTLTISTSGLQASINALGSTATVLTSADVMDENSFSNPTKVV 309
+N + VKV N + ++++ GL+ L S D+ +N+ P +V
Sbjct: 572 KGKNELI-VKVANTSATIQPISLNFEGLKKQDVLSNGRCIKLRSLDLDKDNTLEQPFGIV 630
Query: 308 PVKSDLSNAAEQMQVTLAPRSFSSF 234
P ++ +S L P +F+ +
Sbjct: 631 PQETPVSIEGNVFTTELEPTTFAVY 655
>sptr|Q8AAU6|Q8AAU6 Alpha-L-arabinofuranosidase A precursor.
Length = 660
Score = 44.7 bits (104), Expect = 0.001
Identities = 31/144 (21%), Positives = 59/144 (40%), Gaps = 4/144 (2%)
Frame = -3
Query: 653 RWNPDAIVFNSWQHYGTPSYWMQTLFRESSGAMFHPTTI----XXXXXXXXXXXAITWQD 486
+W PD I F++ T SY++Q L+ ++ G P T+ A D
Sbjct: 512 QWRPDMIWFDNLNSVRTVSYYVQQLYAQNKGTNVLPLTMNKKPVTGAEGQNGLFASAVYD 571
Query: 485 SENSFLRVKVINFGSDAVTLTISTSGLQASINALGSTATVLTSADVMDENSFSNPTKVVP 306
+ + L VKV N ++++ GL+ L+S D +N+ P + P
Sbjct: 572 KDKNELIVKVANTSDKTQPVSLTFEGLKKQDVLSEGRCITLSSLDQDKDNTLEQPFAITP 631
Query: 305 VKSDLSNAAEQMQVTLAPRSFSSF 234
++ ++ + L P +F+ +
Sbjct: 632 QETPVTINGHALTTELGPNTFAVY 655
>sptr|Q8A1K4|Q8A1K4 Alpha-L-arabinofuranosidase A precursor.
Length = 825
Score = 37.7 bits (86), Expect = 0.16
Identities = 25/105 (23%), Positives = 50/105 (47%), Gaps = 3/105 (2%)
Frame = -3
Query: 659 KNRWNPDAIVFNSWQHYGTPSYWMQTLFRESSGAMFHPTTIXXXXXXXXXXXAI---TWQ 489
+ +WNPD I FN+ + T Y++Q L+ +++G + P+ I + +
Sbjct: 682 RTQWNPDLIYFNNREVRPTTGYYVQKLYGQNAGDHYIPSQINLDNQDGRVKLRVGSSIVR 741
Query: 488 DSENSFLRVKVINFGSDAVTLTISTSGLQASINALGSTATVLTSA 354
DS+ + VK++N ++ + G+ ++ +T TVL A
Sbjct: 742 DSKTGDVIVKLVNLLPVSIETDVRLPGMDGIQSS--ATRTVLAGA 784
>sptr|Q8AAW4|Q8AAW4 Alpha-L-arabinofuranosidase.
Length = 514
Score = 34.7 bits (78), Expect = 1.3
Identities = 29/133 (21%), Positives = 58/133 (43%), Gaps = 7/133 (5%)
Frame = -3
Query: 605 TPSYWMQTLFRESSGAMFHPTTIXXXXXXXXXXXAI-----TWQDSENSFLRVKVINFGS 441
TP+Y++ +++ A + P + + T ++N + + + N +
Sbjct: 385 TPTYYVFKMYKVHQDATYLPLDLTCEKMNVRDNRTVPMVSATASKNKNGVIHISLSNVDA 444
Query: 440 D-AVTLTISTSGLQASINALGSTATVLTSADVMDENSFSNPTKVVPVK-SDLSNAAEQMQ 267
D A +T++ +NA + +LTSA++ D NSF P V P ++ M+
Sbjct: 445 DNAQEITVNLP----DVNAKKAIGEILTSANLTDYNSFEKPNIVKPASFKEVKINKGIMK 500
Query: 266 VTLAPRSFSSFDL 228
V L +S + +L
Sbjct: 501 VKLPAKSIVTLEL 513
>sptr|Q9XL35|Q9XL35 NADH dehydrogenase subunit V.
Length = 605
Score = 34.7 bits (78), Expect = 1.3
Identities = 35/122 (28%), Positives = 50/122 (40%), Gaps = 15/122 (12%)
Frame = +1
Query: 232 SNELKERGASVTCICSAALLRSLF-------TGTTLVGLLKEFSSITS--------ADVR 366
SN LK A++T A+ L SL +GT + L E+ I D
Sbjct: 28 SNNLKNTPATITTTVKASFLTSLIPMTIYIHSGTESLTTLWEWKFIMGFKIPVSLKMDFY 87
Query: 367 TVAVDPRALILAWSPLVEIVRVTASDPKFMTFTLRKLFSESCQVIADAASEPE*LELIVV 546
++ P AL ++WS L ASDP F LF +I AA+ L ++ +
Sbjct: 88 SLTFFPIALFVSWSILQFATWYMASDPYITKFFTYLLFFLMAMLILIAANN---LFVLFI 144
Query: 547 GW 552
GW
Sbjct: 145 GW 146
>sptr|O88043|O88043 Putative secreted arabinosidase.
Length = 824
Score = 33.9 bits (76), Expect = 2.3
Identities = 14/38 (36%), Positives = 24/38 (63%)
Frame = -3
Query: 653 RWNPDAIVFNSWQHYGTPSYWMQTLFRESSGAMFHPTT 540
+W+PD I FN+ +G+ +Y +Q LF ++G P+T
Sbjct: 510 QWSPDLIWFNNHASWGSANYEVQKLFMNNTGDRVVPST 547
>sptr|Q8PL31|Q8PL31 Flagellar protein.
Length = 399
Score = 32.7 bits (73), Expect = 5.0
Identities = 26/105 (24%), Positives = 48/105 (45%), Gaps = 17/105 (16%)
Frame = -3
Query: 470 LRVKVINFGSDAVTLTISTS---------GLQASINALGSTATVLTSADVMDENSFSNPT 318
L+++ + G D +LT+ TS GLQ + A GSTA LT D+
Sbjct: 254 LKLESLKAGQDFTSLTMGTSSATGVTVGAGLQTASAASGSTAVTLTDLDISTFAGSQQAL 313
Query: 317 KVV--------PVKSDLSNAAEQMQVTLAPRSFSSFDLALAQSKL 207
++V ++D+ + T+A S +S +L+ ++S++
Sbjct: 314 EIVDKALTAVNSSRADMGAVQNRFTSTIANLSATSENLSASRSRI 358
>sptr|Q9EXF9|Q9EXF9 Proline-rich protein (Lmo0320 protein).
Length = 399
Score = 32.3 bits (72), Expect = 6.6
Identities = 23/86 (26%), Positives = 40/86 (46%), Gaps = 10/86 (11%)
Frame = -3
Query: 482 ENSFLRVKVINFGSDAVTLTISTSGLQASINAL----GSTATVLTSAD------VMDENS 333
ENSF+R G + +T+S +Q +IN L T T+LT D V ++ +
Sbjct: 116 ENSFVRSGTETGGDKVIKVTVSYDYIQTAINELFLNNNPTGTILTITDQNAINAVQEQIN 175
Query: 332 FSNPTKVVPVKSDLSNAAEQMQVTLA 255
++ + L+NA +Q+ +A
Sbjct: 176 TVTSSEKESFQEQLNNAQQQLDARIA 201
>sptr|Q97DN6|Q97DN6 Probable alpha-arabinofuranosidase.
Length = 835
Score = 32.3 bits (72), Expect = 6.6
Identities = 14/31 (45%), Positives = 20/31 (64%)
Frame = -3
Query: 653 RWNPDAIVFNSWQHYGTPSYWMQTLFRESSG 561
+W+PD I FN +Y TP+Y +Q LF + G
Sbjct: 515 QWSPDMIWFNGNTNYVTPNYNVQKLFSTNLG 545
Database: /db/trembl-ebi/tmp/swall
Posted date: Sep 26, 2003 8:29 PM
Number of letters in database: 406,886,216
Number of sequences in database: 1,272,877
Lambda K H
0.318 0.135 0.401
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 544,896,049
Number of Sequences: 1272877
Number of extensions: 11559495
Number of successful extensions: 27382
Number of sequences better than 10.0: 20
Number of HSP's better than 10.0 without gapping: 26525
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 27353
length of database: 406,886,216
effective HSP length: 119
effective length of database: 255,413,853
effective search space used: 32182145478
frameshift window, decay const: 50, 0.1
T: 12
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)

