BLASTX 2.2.6 [Apr-09-2003]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
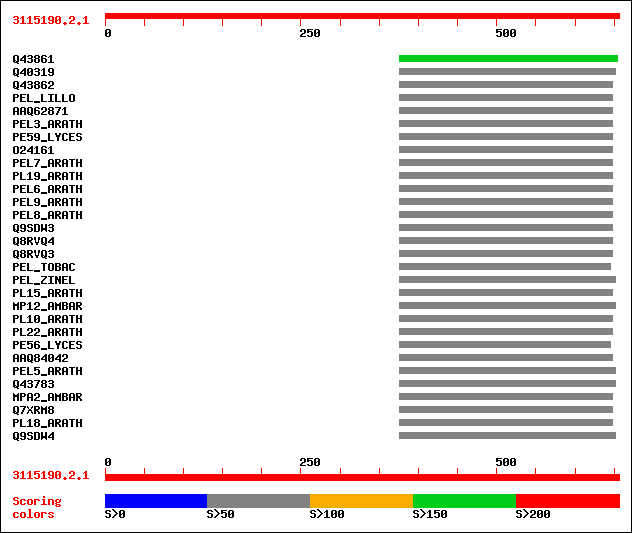
Query= 3115190.2.1
(656 letters)
Database: /db/uniprot/tmp/swall
1,395,590 sequences; 442,889,342 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
sptr|Q43861|Q43861 Pectate lyase homolog (EC 4.2.2.2) (Fragment). 196 2e-49
sptr|Q40319|Q40319 Pectate lyase homolog. 98 8e-20
sptr|Q43862|Q43862 Pectate lyase homolog (EC 4.2.2.2). 94 2e-18
sw|P40973|PEL_LILLO Pectate lyase precursor (EC 4.2.2.2). 92 8e-18
sptrnew|AAQ62871|AAQ62871 At1g14420. 91 1e-17
sw|Q9M9S2|PEL3_ARATH Probable pectate lyase 3 precursor (EC 4.2.... 91 1e-17
sw|P15722|PE59_LYCES Probable pectate lyase P59 precursor (EC 4.... 91 1e-17
sptr|O24161|O24161 Putative pectate lyase Nt59 (Fragment). 87 2e-16
sw|Q9SRH4|PEL7_ARATH Probable pectate lyase 7 precursor (EC 4.2.... 87 2e-16
sw|Q9LFP5|PL19_ARATH Putative pectate lyase 19 precursor (EC 4.2... 85 1e-15
sw|O64510|PEL6_ARATH Probable pectate lyase 6 precursor (EC 4.2.... 84 1e-15
sw|Q9LRM5|PEL9_ARATH Putative pectate lyase 9 precursor (EC 4.2.... 78 9e-14
sw|Q9M8Z8|PEL8_ARATH Probable pectate lyase 8 precursor (EC 4.2.... 78 1e-13
sptr|Q9SDW3|Q9SDW3 Pectate lyase 2. 77 2e-13
sptr|Q8RVQ4|Q8RVQ4 Pectate lyase (Fragment). 77 2e-13
sptr|Q8RVQ3|Q8RVQ3 Pectate lyase (Fragment). 77 2e-13
sw|P40972|PEL_TOBAC Pectate lyase precursor (EC 4.2.2.2). 75 8e-13
sw|O24554|PEL_ZINEL Pectate lyase precursor (EC 4.2.2.2) (ZePel). 74 2e-12
sw|Q944R1|PL15_ARATH Probable pectate lyase 15 precursor (EC 4.2... 74 2e-12
sw|P27760|MP12_AMBAR Pollen allergen Amb a 1.2 precursor (Antige... 74 2e-12
sw|Q9LJ42|PL10_ARATH Probable pectate lyase 10 precursor (EC 4.2... 74 2e-12
sw|Q93Z25|PL22_ARATH Probable pectate lyase 22 precursor (EC 4.2... 73 3e-12
sw|P15721|PE56_LYCES Probable pectate lyase P56 precursor (EC 4.... 73 3e-12
sptrnew|AAQ84042|AAQ84042 Pectate lyase. 73 4e-12
sw|Q9FXD8|PEL5_ARATH Probable pectate lyase 5 precursor (EC 4.2.... 72 5e-12
sptr|Q43783|Q43783 Pectate lyase (EC 4.2.2.2) (Fragment). 72 6e-12
sw|P27762|MPA2_AMBAR Pollen allergen Amb a 2 precursor (Antigen ... 72 8e-12
sptr|Q7XRM8|Q7XRM8 OSJNBa0095E20.8 protein. 72 8e-12
sw|Q9C5M8|PL18_ARATH Probable pectate lyase 18 precursor (EC 4.2... 71 1e-11
sptr|Q9SDW4|Q9SDW4 Pectate lyase 1. 70 3e-11
>sptr|Q43861|Q43861 Pectate lyase homolog (EC 4.2.2.2) (Fragment).
Length = 104
Score = 196 bits (498), Expect = 2e-49
Identities = 89/93 (95%), Positives = 90/93 (96%)
Frame = -3
Query: 654 DAPTIISQGNRFIAPPNIAAKVIPKPYAEEGVWKNWVWHTEDDLFMNGAIFNPSGGAPKQ 475
DAPTIISQGNR+IAPPNIAAKVI K YAEEGVWKNWVWHTEDDLFMNGAIFNPSGGAPKQ
Sbjct: 12 DAPTIISQGNRYIAPPNIAAKVITKHYAEEGVWKNWVWHTEDDLFMNGAIFNPSGGAPKQ 71
Query: 474 VDTNEWVKPKPGTYVTRLTRFSGPLSCCTGKPC 376
VDTNEWVKPKPGTYVTRLTRFSG LSCCTGKPC
Sbjct: 72 VDTNEWVKPKPGTYVTRLTRFSGTLSCCTGKPC 104
>sptr|Q40319|Q40319 Pectate lyase homolog.
Length = 450
Score = 98.2 bits (243), Expect = 8e-20
Identities = 50/95 (52%), Positives = 61/95 (64%), Gaps = 3/95 (3%)
Frame = -3
Query: 651 APTIISQGNRFIAPPNIAAKVIP-KPYAEEGVWKNWVWHTEDDLFMNGAIFNPSGGAPKQ 475
APTI+SQGNRFIAP N AAK + + YA E VW W W +E D FMNGA F SG K
Sbjct: 356 APTILSQGNRFIAPHNNAAKTVTHRDYAPESVWSKWQWRSEGDHFMNGATFIQSGPPIKN 415
Query: 474 VDTNE--WVKPKPGTYVTRLTRFSGPLSCCTGKPC 376
+ + +KP+ G+ RLTRFSG L+C G+PC
Sbjct: 416 LPFKKGFLMKPRHGSQANRLTRFSGALNCVVGRPC 450
>sptr|Q43862|Q43862 Pectate lyase homolog (EC 4.2.2.2).
Length = 438
Score = 94.0 bits (232), Expect = 2e-18
Identities = 48/96 (50%), Positives = 63/96 (65%), Gaps = 5/96 (5%)
Frame = -3
Query: 648 PTIISQGNRFIAPPNIAAKVIPK----PYAEEGVWKNWVWHTEDDLFMNGAIFNPSGGA- 484
PTIISQGNRFIAP + AK + K PY + +K WVW ++ D+ MNGA FN SGG
Sbjct: 346 PTIISQGNRFIAPDDPNAKEVTKREYTPYKD---YKEWVWKSQGDVMMNGAFFNESGGQN 402
Query: 483 PKQVDTNEWVKPKPGTYVTRLTRFSGPLSCCTGKPC 376
++ D +++ K G YV +LTRF+GPL C G+PC
Sbjct: 403 ERKYDRFDFIPAKHGRYVGQLTRFAGPLKCIVGQPC 438
>sw|P40973|PEL_LILLO Pectate lyase precursor (EC 4.2.2.2).
Length = 434
Score = 91.7 bits (226), Expect = 8e-18
Identities = 47/93 (50%), Positives = 58/93 (62%), Gaps = 2/93 (2%)
Frame = -3
Query: 648 PTIISQGNRFIAPPNIAAKVIPK-PYAEEGVWKNWVWHTEDDLFMNGAIFNPSGGA-PKQ 475
PTIISQGNR+IAP AAK + K YAE W W W ++ DLF++GA F SGG +
Sbjct: 342 PTIISQGNRYIAPHIEAAKEVTKRDYAEPAEWSKWTWKSQGDLFVSGAFFVESGGPFENK 401
Query: 474 VDTNEWVKPKPGTYVTRLTRFSGPLSCCTGKPC 376
+ +K KPGT+V RLTRFSG L+C C
Sbjct: 402 YSKKDLIKAKPGTFVQRLTRFSGALNCKENMEC 434
>sptrnew|AAQ62871|AAQ62871 At1g14420.
Length = 459
Score = 91.3 bits (225), Expect = 1e-17
Identities = 51/99 (51%), Positives = 61/99 (61%), Gaps = 8/99 (8%)
Frame = -3
Query: 648 PTIISQGNRFIAPPNIAAKVIPK-PYAEEGVWKNWVWHTEDDLFMNGAIFNPSGGA---- 484
PTIISQGNRFIAPPN AK I K Y G WK+W W +E D F+NGA F SG A
Sbjct: 361 PTIISQGNRFIAPPNEEAKQITKREYTPYGEWKSWNWQSEGDYFLNGAYFVQSGKANAWS 420
Query: 483 --PKQVDTNEW-VKPKPGTYVTRLTRFSGPLSCCTGKPC 376
PK N++ ++PKPGT V +LT +G L C G+ C
Sbjct: 421 SKPKTPLPNKFTIRPKPGTMVRKLTMDAGVLGCKLGEAC 459
>sw|Q9M9S2|PEL3_ARATH Probable pectate lyase 3 precursor (EC
4.2.2.2) (Pectate lyase A2).
Length = 459
Score = 91.3 bits (225), Expect = 1e-17
Identities = 51/99 (51%), Positives = 61/99 (61%), Gaps = 8/99 (8%)
Frame = -3
Query: 648 PTIISQGNRFIAPPNIAAKVIPK-PYAEEGVWKNWVWHTEDDLFMNGAIFNPSGGA---- 484
PTIISQGNRFIAPPN AK I K Y G WK+W W +E D F+NGA F SG A
Sbjct: 361 PTIISQGNRFIAPPNEEAKQITKREYTPYGEWKSWNWQSEGDYFLNGAYFVQSGKANAWS 420
Query: 483 --PKQVDTNEW-VKPKPGTYVTRLTRFSGPLSCCTGKPC 376
PK N++ ++PKPGT V +LT +G L C G+ C
Sbjct: 421 SKPKTPLPNKFTIRPKPGTMVRKLTMDAGVLGCKLGEAC 459
>sw|P15722|PE59_LYCES Probable pectate lyase P59 precursor (EC
4.2.2.2).
Length = 449
Score = 90.9 bits (224), Expect = 1e-17
Identities = 47/95 (49%), Positives = 56/95 (58%), Gaps = 4/95 (4%)
Frame = -3
Query: 648 PTIISQGNRFIAPPNIAAKVIPK-PYAEEGVWKNWVWHTEDDLFMNGAIFNPSGG---AP 481
PTII QGNRFIAPP+I K + K Y E VW W W +E +LFMNGA F SG +
Sbjct: 355 PTIIHQGNRFIAPPDIFKKQVTKREYNPESVWMQWTWRSEGNLFMNGAYFTESGDPEWSS 414
Query: 480 KQVDTNEWVKPKPGTYVTRLTRFSGPLSCCTGKPC 376
K D + + P VT +TRF+G L C GKPC
Sbjct: 415 KHKDLYDGISAAPAEDVTWMTRFAGVLGCKPGKPC 449
>sptr|O24161|O24161 Putative pectate lyase Nt59 (Fragment).
Length = 171
Score = 87.0 bits (214), Expect = 2e-16
Identities = 45/95 (47%), Positives = 58/95 (61%), Gaps = 4/95 (4%)
Frame = -3
Query: 648 PTIISQGNRFIAPPNIAAKVIPK-PYAEEGVWKNWVWHTEDDLFMNGAIFNPSGG---AP 481
PTII+QGNRFIAPP+I + + K Y E VWK+W W +E +LFMNGA F SG +
Sbjct: 77 PTIITQGNRFIAPPDIFKEQVTKREYNPEEVWKHWTWRSEGNLFMNGAYFIESGDPDWSK 136
Query: 480 KQVDTNEWVKPKPGTYVTRLTRFSGPLSCCTGKPC 376
K + + + P VT +TRF+G L C GK C
Sbjct: 137 KHKELYDGISAAPAEEVTWITRFAGALGCKKGKAC 171
>sw|Q9SRH4|PEL7_ARATH Probable pectate lyase 7 precursor (EC
4.2.2.2).
Length = 475
Score = 87.0 bits (214), Expect = 2e-16
Identities = 43/95 (45%), Positives = 57/95 (60%), Gaps = 4/95 (4%)
Frame = -3
Query: 648 PTIISQGNRFIAPPNIA--AKVIPKPYAEEGVWKNWVWHTEDDLFMNGAIFNPSGGAPKQ 475
PTI+S GNRFIAPP+ +V + YA E WKNW W +E D+FMN A F SG +
Sbjct: 381 PTILSHGNRFIAPPHKQHYREVTKRDYASESEWKNWNWRSEKDVFMNNAYFRQSGNPHFK 440
Query: 474 V--DTNEWVKPKPGTYVTRLTRFSGPLSCCTGKPC 376
+ +KPK G V++LT+++G L C GK C
Sbjct: 441 CSHSRQQMIKPKNGMAVSKLTKYAGALDCRVGKAC 475
>sw|Q9LFP5|PL19_ARATH Putative pectate lyase 19 precursor (EC
4.2.2.2).
Length = 472
Score = 84.7 bits (208), Expect = 1e-15
Identities = 41/95 (43%), Positives = 58/95 (61%), Gaps = 4/95 (4%)
Frame = -3
Query: 648 PTIISQGNRFIAPPNIA--AKVIPKPYAEEGVWKNWVWHTEDDLFMNGAIFNPSGGAPKQ 475
PTI+S GNRFIAPP+ +V + YA E WK+W W ++ D+FMNGA F SG +
Sbjct: 378 PTILSHGNRFIAPPHKPHYREVTKRDYASEDEWKHWNWRSDKDVFMNGAYFRQSGNPQYK 437
Query: 474 V--DTNEWVKPKPGTYVTRLTRFSGPLSCCTGKPC 376
+ +KPK G V++LT+++G L C G+ C
Sbjct: 438 CAHTRQQMIKPKNGLAVSKLTKYAGALDCRVGRRC 472
>sw|O64510|PEL6_ARATH Probable pectate lyase 6 precursor (EC
4.2.2.2).
Length = 455
Score = 84.3 bits (207), Expect = 1e-15
Identities = 49/102 (48%), Positives = 61/102 (59%), Gaps = 11/102 (10%)
Frame = -3
Query: 648 PTIISQGNRFIAPPNIAAKVIPK----PYAEEGVWKNWVWHTEDDLFMNGAIFNPSGGA- 484
PTIISQGNRFIAPP +K + K PY E WK+W W +E D F+NGA F SG A
Sbjct: 357 PTIISQGNRFIAPPIEDSKQVTKREYTPYPE---WKSWNWQSEKDYFLNGAYFVQSGKAN 413
Query: 483 -----PKQVDTNEW-VKPKPGTYVTRLTRFSGPLSCCTGKPC 376
PK ++ ++P+PGT V RLT+ +G L C GK C
Sbjct: 414 AWSATPKNPIPRKFAIRPQPGTKVRRLTKDAGTLGCKPGKSC 455
>sw|Q9LRM5|PEL9_ARATH Putative pectate lyase 9 precursor (EC
4.2.2.2).
Length = 452
Score = 78.2 bits (191), Expect = 9e-14
Identities = 41/92 (44%), Positives = 50/92 (54%), Gaps = 1/92 (1%)
Frame = -3
Query: 648 PTIISQGNRFIAPPNIAAKVIPKPYAEEG-VWKNWVWHTEDDLFMNGAIFNPSGGAPKQV 472
PTI SQGNRF AP N +AK + K +G W W W +E DL +NGA F PSG
Sbjct: 360 PTINSQGNRFAAPKNHSAKEVTKRLDTKGNEWMEWNWRSEKDLLVNGAFFTPSGEGASGD 419
Query: 471 DTNEWVKPKPGTYVTRLTRFSGPLSCCTGKPC 376
+ KP + V +T +G LSC GKPC
Sbjct: 420 SQTLSLPAKPASMVDAITASAGALSCRRGKPC 451
>sw|Q9M8Z8|PEL8_ARATH Probable pectate lyase 8 precursor (EC
4.2.2.2).
Length = 416
Score = 77.8 bits (190), Expect = 1e-13
Identities = 42/93 (45%), Positives = 53/93 (56%), Gaps = 2/93 (2%)
Frame = -3
Query: 648 PTIISQGNRFIAPPNIAAKVIPK-PYAEEGVWKNWVWHTEDDLFMNGAIFNPSG-GAPKQ 475
PTI SQGNRF+AP N AK + K Y E WK+W W +E DLF+NGA F SG GA
Sbjct: 324 PTINSQGNRFLAPVNPFAKEVTKREYTGESKWKHWNWRSEGDLFLNGAFFTRSGAGAGAN 383
Query: 474 VDTNEWVKPKPGTYVTRLTRFSGPLSCCTGKPC 376
+ K + V +T +SG L+C G+ C
Sbjct: 384 YARASSLSAKSSSLVGTMTSYSGALNCRAGRRC 416
>sptr|Q9SDW3|Q9SDW3 Pectate lyase 2.
Length = 454
Score = 77.0 bits (188), Expect = 2e-13
Identities = 42/93 (45%), Positives = 51/93 (54%), Gaps = 2/93 (2%)
Frame = -3
Query: 648 PTIISQGNRFIAPPNIAAKVIPKPY-AEEGVWKNWVWHTEDDLFMNGAIFNPSG-GAPKQ 475
PTI SQGNR++AP N AK + K ++ WKNW W +E DL +NGA F PSG GA
Sbjct: 362 PTINSQGNRYLAPTNPFAKEVTKRVDTDQSTWKNWNWRSEGDLLLNGAFFTPSGAGASAS 421
Query: 474 VDTNEWVKPKPGTYVTRLTRFSGPLSCCTGKPC 376
KP + V LT +G LSC G C
Sbjct: 422 YARASSFGAKPSSLVDTLTSDAGVLSCQVGTRC 454
>sptr|Q8RVQ4|Q8RVQ4 Pectate lyase (Fragment).
Length = 99
Score = 77.0 bits (188), Expect = 2e-13
Identities = 42/93 (45%), Positives = 51/93 (54%), Gaps = 2/93 (2%)
Frame = -3
Query: 648 PTIISQGNRFIAPPNIAAKVIPKPY-AEEGVWKNWVWHTEDDLFMNGAIFNPSG-GAPKQ 475
PTI SQGNR++AP N AK + K ++ WKNW W +E DL +NGA F PSG GA
Sbjct: 7 PTINSQGNRYLAPTNPFAKEVTKRVDTDQSTWKNWNWRSEGDLLLNGAFFTPSGAGASAS 66
Query: 474 VDTNEWVKPKPGTYVTRLTRFSGPLSCCTGKPC 376
KP + V LT +G LSC G C
Sbjct: 67 YARASSFGAKPSSLVDTLTSDAGVLSCQVGTRC 99
>sptr|Q8RVQ3|Q8RVQ3 Pectate lyase (Fragment).
Length = 122
Score = 77.0 bits (188), Expect = 2e-13
Identities = 42/93 (45%), Positives = 51/93 (54%), Gaps = 2/93 (2%)
Frame = -3
Query: 648 PTIISQGNRFIAPPNIAAKVIPKPY-AEEGVWKNWVWHTEDDLFMNGAIFNPSG-GAPKQ 475
PTI SQGNR++AP N AK + K ++ WKNW W +E DL +NGA F PSG GA
Sbjct: 30 PTINSQGNRYLAPTNPFAKEVTKRVDTDQSTWKNWNWRSEGDLLLNGAFFTPSGAGASAS 89
Query: 474 VDTNEWVKPKPGTYVTRLTRFSGPLSCCTGKPC 376
KP + V LT +G LSC G C
Sbjct: 90 YARASSFGAKPSSLVDTLTSDAGVLSCQVGTRC 122
>sw|P40972|PEL_TOBAC Pectate lyase precursor (EC 4.2.2.2).
Length = 397
Score = 75.1 bits (183), Expect = 8e-13
Identities = 41/95 (43%), Positives = 52/95 (54%), Gaps = 5/95 (5%)
Frame = -3
Query: 645 TIISQGNRFIAPPNIAAKVIP---KPYAEEGVWKNWVWHTEDDLFMNGAIFNPSGGAP-- 481
TIISQGNRFIA + K + K A W W W ++ D NGA F PSG
Sbjct: 303 TIISQGNRFIAEDELLVKEVTYREKLTASVAEWMKWTWISDGDDMENGATFTPSGDQNLL 362
Query: 480 KQVDTNEWVKPKPGTYVTRLTRFSGPLSCCTGKPC 376
++D +KP+P + V LT+FSG LSC G+PC
Sbjct: 363 DKIDHLNLIKPEPSSKVGILTKFSGALSCVKGRPC 397
>sw|O24554|PEL_ZINEL Pectate lyase precursor (EC 4.2.2.2) (ZePel).
Length = 401
Score = 73.9 bits (180), Expect = 2e-12
Identities = 41/94 (43%), Positives = 50/94 (53%), Gaps = 2/94 (2%)
Frame = -3
Query: 651 APTIISQGNRFIAPPNIAAKVIPK-PYAEEGVWKNWVWHTEDDLFMNGAIFNPSGG-APK 478
+PTI SQGNRF+AP K + K A E WKNW W +E DL +NGA F SGG A
Sbjct: 308 SPTIYSQGNRFLAPNTRFDKEVTKHENAPESEWKNWNWRSEGDLMLNGAYFRESGGRAAS 367
Query: 477 QVDTNEWVKPKPGTYVTRLTRFSGPLSCCTGKPC 376
+ +P T V +TR +G L C G C
Sbjct: 368 SFARASSLSGRPSTLVASMTRSAGALVCRKGSRC 401
>sw|Q944R1|PL15_ARATH Probable pectate lyase 15 precursor (EC
4.2.2.2) (Pectate lyase A11).
Length = 470
Score = 73.6 bits (179), Expect = 2e-12
Identities = 39/93 (41%), Positives = 50/93 (53%), Gaps = 2/93 (2%)
Frame = -3
Query: 648 PTIISQGNRFIAPPNIAAKVIPKPY-AEEGVWKNWVWHTEDDLFMNGAIFNPSG-GAPKQ 475
PTI SQGNR+ AP + AK + K + WK W W +E DL +NGA F PSG GA
Sbjct: 378 PTINSQGNRYAAPMDRFAKEVTKRVETDASEWKKWNWRSEGDLLLNGAFFRPSGAGASAS 437
Query: 474 VDTNEWVKPKPGTYVTRLTRFSGPLSCCTGKPC 376
+ KP + V +T +G L C G+PC
Sbjct: 438 YGRASSLAAKPSSMVDTITSTAGALGCRKGRPC 470
>sw|P27760|MP12_AMBAR Pollen allergen Amb a 1.2 precursor (Antigen
E) (Antigen Amb a I).
Length = 398
Score = 73.6 bits (179), Expect = 2e-12
Identities = 42/95 (44%), Positives = 52/95 (54%), Gaps = 3/95 (3%)
Frame = -3
Query: 651 APTIISQGNRFIAPPNIAAK-VIPKPYAEEGVWKNWVWHTEDDLFMNGAIFNPSGGAPKQ 475
APTI+SQGNRF AP +I K V+ + +W W T+ DL NGAIF PSG P
Sbjct: 304 APTILSQGNRFFAPDDIIKKNVLARTGTGNAESMSWNWRTDRDLLENGAIFLPSGSDPVL 363
Query: 474 VDTNE--WVKPKPGTYVTRLTRFSGPLSCCTGKPC 376
+ + +PG V RLT +G LSC G PC
Sbjct: 364 TPEQKAGMIPAEPGEAVLRLTSSAGVLSCHQGAPC 398
>sw|Q9LJ42|PL10_ARATH Probable pectate lyase 10 precursor (EC
4.2.2.2).
Length = 440
Score = 73.6 bits (179), Expect = 2e-12
Identities = 41/93 (44%), Positives = 50/93 (53%), Gaps = 2/93 (2%)
Frame = -3
Query: 648 PTIISQGNRFIAPPNIAAKVIPKPYAE-EGVWKNWVWHTEDDLFMNGAIFNPSG-GAPKQ 475
PTI SQGNRF+AP N AK + K +G WK W W ++ DL +NGA F SG AP
Sbjct: 348 PTINSQGNRFLAPGNPFAKEVTKRVGSWQGEWKQWNWRSQGDLMLNGAYFTKSGAAAPAS 407
Query: 474 VDTNEWVKPKPGTYVTRLTRFSGPLSCCTGKPC 376
+ KP + V+ LT SG L C G C
Sbjct: 408 YARASSLGAKPASVVSMLTYSSGALKCRIGMRC 440
>sw|Q93Z25|PL22_ARATH Probable pectate lyase 22 precursor (EC
4.2.2.2).
Length = 432
Score = 73.2 bits (178), Expect = 3e-12
Identities = 39/95 (41%), Positives = 54/95 (56%), Gaps = 4/95 (4%)
Frame = -3
Query: 648 PTIISQGNRFIAPPNIAAKVIPKPY-AEEGVWKNWVWHTEDDLFMNGAIFNPSGGAPKQV 472
PTI SQGNRF+AP + ++K + K A E W+NW W +E DL +NGA F SG P +
Sbjct: 338 PTINSQGNRFLAPDDSSSKEVTKHEDAPEDEWRNWNWRSEGDLLLNGAFFTYSGAGPAKS 397
Query: 471 DT---NEWVKPKPGTYVTRLTRFSGPLSCCTGKPC 376
+ + +P ++V +T SG LSC G C
Sbjct: 398 SSYSKASSLAARPSSHVGEITIASGALSCKRGSHC 432
>sw|P15721|PE56_LYCES Probable pectate lyase P56 precursor (EC
4.2.2.2).
Length = 398
Score = 73.2 bits (178), Expect = 3e-12
Identities = 41/97 (42%), Positives = 53/97 (54%), Gaps = 7/97 (7%)
Frame = -3
Query: 645 TIISQGNRFIAPPNIAAKVIPKPYAEEGV-----WKNWVWHTEDDLFMNGAIFNPSG--G 487
TIISQGNRFIA + K + Y E+ W W W T+ D F NGA F PSG
Sbjct: 304 TIISQGNRFIAEDKLLVKEVT--YREKSTSSVEEWMKWTWITDGDDFENGATFTPSGDQN 361
Query: 486 APKQVDTNEWVKPKPGTYVTRLTRFSGPLSCCTGKPC 376
++D ++P+P + V LT+FSG LSC +PC
Sbjct: 362 LLSKIDHLNLIQPEPSSKVGLLTKFSGALSCKIRRPC 398
>sptrnew|AAQ84042|AAQ84042 Pectate lyase.
Length = 418
Score = 72.8 bits (177), Expect = 4e-12
Identities = 42/93 (45%), Positives = 50/93 (53%), Gaps = 2/93 (2%)
Frame = -3
Query: 648 PTIISQGNRFIAPPNIAAKVIPKPY-AEEGVWKNWVWHTEDDLFMNGAIFNPSG-GAPKQ 475
PTI SQGNRF AP ++K + K A E WKNW W +E DL +NGA F SG GA
Sbjct: 326 PTINSQGNRFAAPDIRSSKEVTKHEDAPESEWKNWNWRSEGDLMLNGAFFTASGAGASSS 385
Query: 474 VDTNEWVKPKPGTYVTRLTRFSGPLSCCTGKPC 376
+ KP + V +T SG LSC G C
Sbjct: 386 YARASSLGAKPSSLVGAITTASGALSCRKGSRC 418
>sw|Q9FXD8|PEL5_ARATH Probable pectate lyase 5 precursor (EC
4.2.2.2).
Length = 408
Score = 72.4 bits (176), Expect = 5e-12
Identities = 40/94 (42%), Positives = 50/94 (53%), Gaps = 2/94 (2%)
Frame = -3
Query: 651 APTIISQGNRFIAPPNIAAKVIPK-PYAEEGVWKNWVWHTEDDLFMNGAIFNPS-GGAPK 478
APTI SQGNRF+AP + K + K A WK W W +E DLF+NGA F PS GGA
Sbjct: 315 APTINSQGNRFLAPNDHVFKEVTKYEDAPRSKWKKWNWRSEGDLFLNGAFFTPSGGGASS 374
Query: 477 QVDTNEWVKPKPGTYVTRLTRFSGPLSCCTGKPC 376
+ +P + V +T +G L C G C
Sbjct: 375 SYAKASSLSARPSSLVASVTSNAGALFCRKGSRC 408
>sptr|Q43783|Q43783 Pectate lyase (EC 4.2.2.2) (Fragment).
Length = 398
Score = 72.0 bits (175), Expect = 6e-12
Identities = 41/94 (43%), Positives = 52/94 (55%), Gaps = 2/94 (2%)
Frame = -3
Query: 651 APTIISQGNRFIAPPNIAAKVIPKPY-AEEGVWKNWVWHTEDDLFMNGAIFNPSG-GAPK 478
APTI SQGNRF+AP + AK + K A+E WK W W +E D +NGA F PSG GA
Sbjct: 305 APTINSQGNRFLAPNDRFAKEVTKREDAQESEWKKWNWRSEGDQMLNGAFFTPSGAGASS 364
Query: 477 QVDTNEWVKPKPGTYVTRLTRFSGPLSCCTGKPC 376
+ P+ + V +T +G LSC G C
Sbjct: 365 SHAKASSLGPRSSSLVGTITVSAGVLSCKKGSRC 398
>sw|P27762|MPA2_AMBAR Pollen allergen Amb a 2 precursor (Antigen K)
(Antigen Amb a II).
Length = 397
Score = 71.6 bits (174), Expect = 8e-12
Identities = 42/96 (43%), Positives = 56/96 (58%), Gaps = 5/96 (5%)
Frame = -3
Query: 648 PTIISQGNRFIAPPNIAAK-VIPKPYAEEGVWKNWVWHTEDDLFMNGAIFNPSGGAPKQV 472
PTI+SQGN+F+AP I K V + A+E W W W T++D+ NGAIF SG P V
Sbjct: 304 PTILSQGNKFVAPDFIYKKNVCLRTGAQEPEWMTWNWRTQNDVLENGAIFVASGSDP--V 361
Query: 471 DTNE----WVKPKPGTYVTRLTRFSGPLSCCTGKPC 376
T E ++ +PG V +LT +G L+C G PC
Sbjct: 362 LTAEQNAGMMQAEPGDMVPQLTMNAGVLTCSPGAPC 397
>sptr|Q7XRM8|Q7XRM8 OSJNBa0095E20.8 protein.
Length = 472
Score = 71.6 bits (174), Expect = 8e-12
Identities = 39/93 (41%), Positives = 50/93 (53%), Gaps = 2/93 (2%)
Frame = -3
Query: 648 PTIISQGNRFIAPPNIAAKVIPKPY-AEEGVWKNWVWHTEDDLFMNGAIFNPSG-GAPKQ 475
PTI SQGNR++AP N AK + K + +WK W W +E DL +NGA F PSG GA
Sbjct: 380 PTINSQGNRYLAPTNPFAKEVTKRVETAQTIWKGWNWRSEGDLLLNGAFFTPSGAGASAS 439
Query: 474 VDTNEWVKPKPGTYVTRLTRFSGPLSCCTGKPC 376
+ K + V +T +G LSC G C
Sbjct: 440 YSRASSLGAKSSSMVGTITSGAGALSCRGGSAC 472
>sw|Q9C5M8|PL18_ARATH Probable pectate lyase 18 precursor (EC
4.2.2.2) (Pectate lyase A10).
Length = 408
Score = 70.9 bits (172), Expect = 1e-11
Identities = 41/93 (44%), Positives = 49/93 (52%), Gaps = 2/93 (2%)
Frame = -3
Query: 648 PTIISQGNRFIAPPNIAAKVIPKPY-AEEGVWKNWVWHTEDDLFMNGAIFNPSGGAPKQV 472
PTI SQGNRF+AP +K + K A E WK W W + DL +NGA F PSGGA
Sbjct: 316 PTINSQGNRFLAPNIRFSKEVTKHEDAPESEWKRWNWRSSGDLLLNGAFFTPSGGAASSS 375
Query: 471 DTN-EWVKPKPGTYVTRLTRFSGPLSCCTGKPC 376
+ KP + V LT SG L+C G C
Sbjct: 376 YAKASSLGAKPSSLVGPLTSTSGALNCRKGSRC 408
>sptr|Q9SDW4|Q9SDW4 Pectate lyase 1.
Length = 407
Score = 69.7 bits (169), Expect = 3e-11
Identities = 40/94 (42%), Positives = 51/94 (54%), Gaps = 2/94 (2%)
Frame = -3
Query: 651 APTIISQGNRFIAPPNIAAKVIPKPY-AEEGVWKNWVWHTEDDLFMNGAIFNPSG-GAPK 478
APTI SQGNRF+AP + AK + K A+E WK W W +E D +NGA F PSG GA
Sbjct: 314 APTINSQGNRFLAPNDRFAKEVTKREDAQESEWKKWNWRSEGDQMLNGAFFTPSGAGASS 373
Query: 477 QVDTNEWVKPKPGTYVTRLTRFSGPLSCCTGKPC 376
+ + + V +T +G LSC G C
Sbjct: 374 SYAKASSLGARSSSLVGTITVSAGVLSCKKGSRC 407
Database: /db/uniprot/tmp/swall
Posted date: Mar 5, 2004 7:45 PM
Number of letters in database: 442,889,342
Number of sequences in database: 1,395,590
Lambda K H
0.318 0.135 0.401
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 543,170,520
Number of Sequences: 1395590
Number of extensions: 12217175
Number of successful extensions: 38139
Number of sequences better than 10.0: 30
Number of HSP's better than 10.0 without gapping: 36844
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 38010
length of database: 442,889,342
effective HSP length: 118
effective length of database: 278,209,722
effective search space used: 27820972200
frameshift window, decay const: 50, 0.1
T: 12
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)

