BLASTX 2.2.6 [Apr-09-2003]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= 3023930.2.1
(544 letters)
Database: /db/trembl-ebi/tmp/swall
1,272,877 sequences; 406,886,216 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
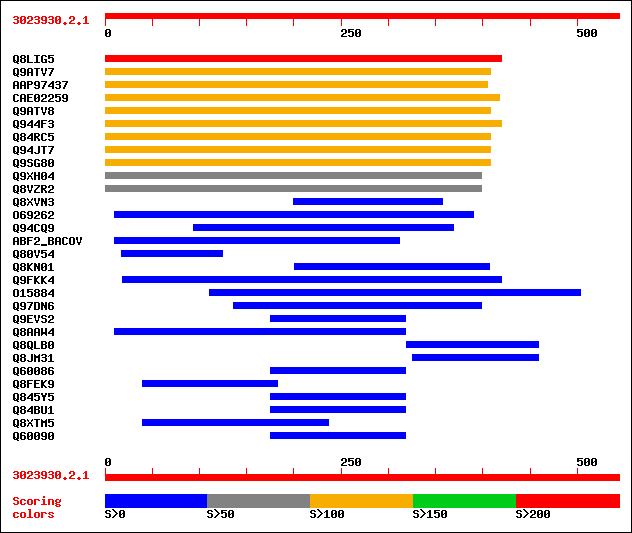
sptr|Q8LIG5|Q8LIG5 Putative arabinoxylan narabinofuranohydrolase... 226 1e-58
sptr|Q9ATV7|Q9ATV7 Arabinoxylan arabinofuranohydrolase isoenzyme... 133 1e-30
sptrnew|AAP97437|AAP97437 Alpha-L-arabinofuranosidase. 131 5e-30
sptrnew|CAE02259|CAE02259 OSJNBb0058J09.6 protein. 128 5e-29
sptr|Q9ATV8|Q9ATV8 Arabinoxylan arabinofuranohydrolase isoenzyme... 126 1e-28
sptr|Q944F3|Q944F3 Arabinosidase ARA-1. 125 4e-28
sptr|Q84RC5|Q84RC5 Alpha-L-arabinofuranosidase. 113 1e-24
sptr|Q94JT7|Q94JT7 AT3g10740/T7M13_18. 113 1e-24
sptr|Q9SG80|Q9SG80 Putative alpha-L-arabinofuranosidase. 113 1e-24
sptr|Q9XH04|Q9XH04 T1N24.13 protein. 97 1e-19
sptr|Q8VZR2|Q8VZR2 Putative arabinosidase (Alpha-L-arabinofurano... 97 1e-19
sptr|Q8XVN3|Q8XVN3 Putative hemagglutinin-related transmembrane ... 35 0.41
sptr|O69262|O69262 Alpha-L-arabinofuranosidase (EC 3.2.1.55). 35 0.70
sptr|Q94CQ9|Q94CQ9 Putative beta 1,3-glucanase. 33 1.6
sw|Q59219|ABF2_BACOV Alpha-L-arabinofuranosidase 2 (EC 3.2.1.55)... 33 1.6
sptr|Q80V54|Q80V54 1300015B04Rik protein. 33 2.0
sptr|Q8KN01|Q8KN01 Hypothetical protein Ste20. 33 2.0
sptr|Q9FKK4|Q9FKK4 Genomic DNA, chromosome 5, P1 clone:MUF9 (AT5... 33 2.6
sptr|O15884|O15884 Invariant surface glycoprotein 100. 33 2.6
sptr|Q97DN6|Q97DN6 Probable alpha-arabinofuranosidase. 33 2.6
sptr|Q9EVS2|Q9EVS2 FtsZ protein (Cell division protein ftsZ) (Fr... 32 3.5
sptr|Q8AAW4|Q8AAW4 Alpha-L-arabinofuranosidase. 32 3.5
sptr|Q8QLB0|Q8QLB0 LEF-9. 32 3.5
sptr|Q8JM31|Q8JM31 Putative late expression factor LEF-9. 32 4.5
sptr|Q60086|Q60086 Cell division protein ftsZ (Fragment). 32 4.5
sptr|Q8FEK9|Q8FEK9 Hypothetical protein ygbK. 32 4.5
sptr|Q845Y5|Q845Y5 Cell division protein (Fragment). 32 5.9
sptr|Q84BU1|Q84BU1 Cell division protein (Fragment). 32 5.9
sptr|Q8XTM5|Q8XTM5 Hypothetical protein RSp0081. 32 5.9
sptr|Q60090|Q60090 Cell division protein ftsZ (Fragment). 32 5.9
>sptr|Q8LIG5|Q8LIG5 Putative arabinoxylan narabinofuranohydrolase
isoenzyme AXAH-I.
Length = 664
Score = 226 bits (576), Expect = 1e-58
Identities = 107/140 (76%), Positives = 125/140 (89%)
Frame = +1
Query: 1 YGCPNYWMLHFFKESSGAILHPTAIQVSSYDQLVASAITWQNAKDKSTYLRIKVVNFGNQ 180
YGCPNYWMLHFFK+S GA HP+ IQ+SSY+QLVASAITWQN+KDKSTYL+IK+VNFGNQ
Sbjct: 525 YGCPNYWMLHFFKDSCGATFHPSNIQISSYNQLVASAITWQNSKDKSTYLKIKLVNFGNQ 584
Query: 181 AVDLNISVAELATGVKKSGSKQTVLTSSSPLDENSFQQPEKVAPVSSPMANAGQQMGASV 360
AV+L+ISV+ L G+K SGSK+TVLTSS PLDENSFQQP+KVAPVSSP+ NA +QMG V
Sbjct: 585 AVNLSISVSGLDEGIKSSGSKKTVLTSSGPLDENSFQQPQKVAPVSSPVDNANEQMGVLV 644
Query: 361 GPYSLTSFDLLLEPSKHDSV 420
PYSLTSFDLLL+PSKH ++
Sbjct: 645 DPYSLTSFDLLLQPSKHSTI 664
>sptr|Q9ATV7|Q9ATV7 Arabinoxylan arabinofuranohydrolase isoenzyme
AXAH-II.
Length = 656
Score = 133 bits (335), Expect = 1e-30
Identities = 68/136 (50%), Positives = 90/136 (66%)
Frame = +1
Query: 1 YGCPNYWMLHFFKESSGAILHPTAIQVSSYDQLVASAITWQNAKDKSTYLRIKVVNFGNQ 180
YG P+YWM FF+ESSGA++HP I S L ASAITWQ++ + +LR+K+VNFG+
Sbjct: 518 YGTPSYWMQKFFRESSGAMIHPITISSSYSGSLAASAITWQDSGNN--FLRVKIVNFGSD 575
Query: 181 AVDLNISVAELATGVKKSGSKQTVLTSSSPLDENSFQQPEKVAPVSSPMANAGQQMGASV 360
V L ISV+ L + GS TVLTSS+ DENSF P KV PV+S + NA +QM ++
Sbjct: 576 TVSLTISVSGLQASINALGSNATVLTSSNVKDENSFSNPTKVVPVTSQLHNAAEQMQVTL 635
Query: 361 GPYSLTSFDLLLEPSK 408
+S +SFDL L S+
Sbjct: 636 AAHSFSSFDLALAQSE 651
>sptrnew|AAP97437|AAP97437 Alpha-L-arabinofuranosidase.
Length = 675
Score = 131 bits (329), Expect = 5e-30
Identities = 67/136 (49%), Positives = 95/136 (69%), Gaps = 1/136 (0%)
Frame = +1
Query: 1 YGCPNYWMLHFFKESSGAILHPTAIQVSSYDQLVASAITWQNAKDKSTYLRIKVVNFGNQ 180
YG P+YW+ F ESSGA L+ + +Q++S L+ASAI+WQN+++++TYLRIKVVN G
Sbjct: 524 YGTPSYWVQCLFNESSGATLYNSTLQMNSSTSLLASAISWQNSENENTYLRIKVVNLGTY 583
Query: 181 AVDLNISVAEL-ATGVKKSGSKQTVLTSSSPLDENSFQQPEKVAPVSSPMANAGQQMGAS 357
V+L + V L V SGS +TVLTS++ +DENSF +P+KV P S + +AG++M
Sbjct: 584 TVNLKVFVDGLEPNSVSLSGSTKTVLTSNNQMDENSFNEPKKVIPKRSLLESAGEEMEVV 643
Query: 358 VGPYSLTSFDLLLEPS 405
+ P S TS DLL+E S
Sbjct: 644 ISPRSFTSIDLLMESS 659
>sptrnew|CAE02259|CAE02259 OSJNBb0058J09.6 protein.
Length = 790
Score = 128 bits (321), Expect = 5e-29
Identities = 67/139 (48%), Positives = 88/139 (63%)
Frame = +1
Query: 1 YGCPNYWMLHFFKESSGAILHPTAIQVSSYDQLVASAITWQNAKDKSTYLRIKVVNFGNQ 180
YG P+YWM F+ESSGA+LHP I + L ASAITW+ + S++LR+K+VN G+
Sbjct: 654 YGSPSYWMQTIFRESSGAVLHPVTINSMYSNSLAASAITWKASN--SSFLRVKIVNIGSN 711
Query: 181 AVDLNISVAELATGVKKSGSKQTVLTSSSPLDENSFQQPEKVAPVSSPMANAGQQMGASV 360
V+L +S L V S T+LTS + DENSF +P V PV+ + NAG++M A +
Sbjct: 712 PVNLIVSTTGLEALVNMRKSTITILTSKNLSDENSFSKPTNVVPVTRELPNAGEEMFAFL 771
Query: 361 GPYSLTSFDLLLEPSKHDS 417
GPYS TSFDL L KH S
Sbjct: 772 GPYSFTSFDLALGQQKHVS 790
>sptr|Q9ATV8|Q9ATV8 Arabinoxylan arabinofuranohydrolase isoenzyme
AXAH-I.
Length = 658
Score = 126 bits (317), Expect = 1e-28
Identities = 64/136 (47%), Positives = 88/136 (64%)
Frame = +1
Query: 1 YGCPNYWMLHFFKESSGAILHPTAIQVSSYDQLVASAITWQNAKDKSTYLRIKVVNFGNQ 180
YG P+YWM F+ESSGA +HP ++ S L ASAITW + D++++LR+K+VNFG
Sbjct: 520 YGTPSYWMQTLFRESSGATVHPISLSSSCSGLLAASAITWHD--DENSFLRVKIVNFGPD 577
Query: 181 AVDLNISVAELATGVKKSGSKQTVLTSSSPLDENSFQQPEKVAPVSSPMANAGQQMGASV 360
AV L IS L + GS TVLTS +DENSF P KV PV+ + +A ++M ++
Sbjct: 578 AVGLTISATGLQGSINAFGSTATVLTSGGVMDENSFANPNKVVPVTVELRDAAEEMQVTL 637
Query: 361 GPYSLTSFDLLLEPSK 408
P+SLT+FDL L S+
Sbjct: 638 PPHSLTAFDLALAQSR 653
>sptr|Q944F3|Q944F3 Arabinosidase ARA-1.
Length = 674
Score = 125 bits (313), Expect = 4e-28
Identities = 65/142 (45%), Positives = 91/142 (64%), Gaps = 2/142 (1%)
Frame = +1
Query: 1 YGCPNYWMLHFFKESSGAILHPTAIQVSSYDQLVASAITWQNAKDKSTYLRIKVVNFGNQ 180
YG P+YWM HFFKES+GA L +++Q + + L+ASAITW+N+ D + YLRIKVVNFG
Sbjct: 523 YGTPSYWMQHFFKESNGATLLSSSLQANPSNSLIASAITWRNSLDNNDYLRIKVVNFGTT 582
Query: 181 AVDLNISVAELATGVKKS--GSKQTVLTSSSPLDENSFQQPEKVAPVSSPMANAGQQMGA 354
AV IS+ L ++ G+ T LTS++ +DENSF++P KV PV + + M
Sbjct: 583 AVITKISLTGLGQNSLETLFGAVMTELTSNNVMDENSFREPNKVIPVKTQVEKVSDNMDV 642
Query: 355 SVGPYSLTSFDLLLEPSKHDSV 420
+ P SL S D LL S +++V
Sbjct: 643 VLAPRSLNSIDFLLRKSINNNV 664
>sptr|Q84RC5|Q84RC5 Alpha-L-arabinofuranosidase.
Length = 678
Score = 113 bits (283), Expect = 1e-24
Identities = 65/137 (47%), Positives = 90/137 (65%), Gaps = 1/137 (0%)
Frame = +1
Query: 1 YGCPNYWMLHFFKESSGAILHPTAIQVSSYDQLVASAITWQNAKDKSTYLRIKVVNFGNQ 180
YG P+YW+ FF ESSGA L + ++ +S LVASAI+W+N + Y+RIK VNFG
Sbjct: 528 YGTPSYWVQRFFAESSGATLLTSTLKGNS-TSLVASAISWKN--NGKDYIRIKAVNFGAN 584
Query: 181 AVDLNISVAELATGVKK-SGSKQTVLTSSSPLDENSFQQPEKVAPVSSPMANAGQQMGAS 357
+ ++ + V L V + SGSK+TVLTS++ +DENSF QPEKV P S + A + M
Sbjct: 585 SENMQVLVTGLDPNVMRVSGSKKTVLTSTNVMDENSFSQPEKVVPHESLLELAEEDMTVV 644
Query: 358 VGPYSLTSFDLLLEPSK 408
+ P+S +SFDLL E +K
Sbjct: 645 LPPHSFSSFDLLKESAK 661
>sptr|Q94JT7|Q94JT7 AT3g10740/T7M13_18.
Length = 678
Score = 113 bits (283), Expect = 1e-24
Identities = 65/137 (47%), Positives = 90/137 (65%), Gaps = 1/137 (0%)
Frame = +1
Query: 1 YGCPNYWMLHFFKESSGAILHPTAIQVSSYDQLVASAITWQNAKDKSTYLRIKVVNFGNQ 180
YG P+YW+ FF ESSGA L + ++ +S LVASAI+W+N + Y+RIK VNFG
Sbjct: 528 YGTPSYWVQRFFAESSGATLLTSTLKGNS-TSLVASAISWKN--NGKDYIRIKAVNFGAN 584
Query: 181 AVDLNISVAELATGVKK-SGSKQTVLTSSSPLDENSFQQPEKVAPVSSPMANAGQQMGAS 357
+ ++ + V L V + SGSK+TVLTS++ +DENSF QPEKV P S + A + M
Sbjct: 585 SENMQVLVTGLDPNVMRVSGSKKTVLTSTNVMDENSFSQPEKVVPHESLLELAEEDMTVV 644
Query: 358 VGPYSLTSFDLLLEPSK 408
+ P+S +SFDLL E +K
Sbjct: 645 LPPHSFSSFDLLKESAK 661
>sptr|Q9SG80|Q9SG80 Putative alpha-L-arabinofuranosidase.
Length = 678
Score = 113 bits (283), Expect = 1e-24
Identities = 65/137 (47%), Positives = 90/137 (65%), Gaps = 1/137 (0%)
Frame = +1
Query: 1 YGCPNYWMLHFFKESSGAILHPTAIQVSSYDQLVASAITWQNAKDKSTYLRIKVVNFGNQ 180
YG P+YW+ FF ESSGA L + ++ +S LVASAI+W+N + Y+RIK VNFG
Sbjct: 528 YGTPSYWVQRFFAESSGATLLTSTLKGNS-TSLVASAISWKN--NGKDYIRIKAVNFGAN 584
Query: 181 AVDLNISVAELATGVKK-SGSKQTVLTSSSPLDENSFQQPEKVAPVSSPMANAGQQMGAS 357
+ ++ + V L V + SGSK+TVLTS++ +DENSF QPEKV P S + A + M
Sbjct: 585 SENMQVLVTGLDPNVMRVSGSKKTVLTSTNVMDENSFSQPEKVVPHESLLELAEEDMTVV 644
Query: 358 VGPYSLTSFDLLLEPSK 408
+ P+S +SFDLL E +K
Sbjct: 645 LPPHSFSSFDLLKESAK 661
>sptr|Q9XH04|Q9XH04 T1N24.13 protein.
Length = 521
Score = 96.7 bits (239), Expect = 1e-19
Identities = 60/135 (44%), Positives = 85/135 (62%), Gaps = 2/135 (1%)
Frame = +1
Query: 1 YGCPNYWMLHFFKESSGAILHPTAIQVSSYDQLVASAITWQ-NAKDKSTYLRIKVVNFGN 177
YG P+YW+ HFF ESSGA L + ++ + + ASAI++Q N KD Y++IK VNFG
Sbjct: 374 YGTPSYWVQHFFTESSGATLLNSTLKGKT-SSVEASAISFQTNGKD---YIQIKAVNFGE 429
Query: 178 QAVDLNISVAELATGVKKSGSKQTVLTSSSPLDENSFQQPEKVAPVSSPMANAGQQ-MGA 354
Q+V+L ++V L K GSK+ VLTS+S +DENSF P + P S + Q+ +
Sbjct: 430 QSVNLKVAVTGLMA--KFYGSKKKVLTSASVMDENSFSNPNMIVPQESLLEMTEQEDLMF 487
Query: 355 SVGPYSLTSFDLLLE 399
+ P+S +SFDLL E
Sbjct: 488 VLPPHSFSSFDLLTE 502
>sptr|Q8VZR2|Q8VZR2 Putative arabinosidase
(Alpha-L-arabinofuranosidase) (Hypothetical protein
At5g26120).
Length = 674
Score = 96.7 bits (239), Expect = 1e-19
Identities = 60/135 (44%), Positives = 85/135 (62%), Gaps = 2/135 (1%)
Frame = +1
Query: 1 YGCPNYWMLHFFKESSGAILHPTAIQVSSYDQLVASAITWQ-NAKDKSTYLRIKVVNFGN 177
YG P+YW+ HFF ESSGA L + ++ + + ASAI++Q N KD Y++IK VNFG
Sbjct: 527 YGTPSYWVQHFFTESSGATLLNSTLKGKT-SSVEASAISFQTNGKD---YIQIKAVNFGE 582
Query: 178 QAVDLNISVAELATGVKKSGSKQTVLTSSSPLDENSFQQPEKVAPVSSPMANAGQQ-MGA 354
Q+V+L ++V L K GSK+ VLTS+S +DENSF P + P S + Q+ +
Sbjct: 583 QSVNLKVAVTGLMA--KFYGSKKKVLTSASVMDENSFSNPNMIVPQESLLEMTEQEDLMF 640
Query: 355 SVGPYSLTSFDLLLE 399
+ P+S +SFDLL E
Sbjct: 641 VLPPHSFSSFDLLTE 655
>sptr|Q8XVN3|Q8XVN3 Putative hemagglutinin-related transmembrane
protein.
Length = 361
Score = 35.4 bits (80), Expect = 0.41
Identities = 17/53 (32%), Positives = 31/53 (58%)
Frame = +1
Query: 199 SVAELATGVKKSGSKQTVLTSSSPLDENSFQQPEKVAPVSSPMANAGQQMGAS 357
+V+ L G++ +G T+ TSS P+ + + Q V P++S A QQ+G++
Sbjct: 173 TVSALGQGLQGAGGLVTIATSSGPVQQVTQQTSNAVVPLTSQTARGTQQVGST 225
>sptr|O69262|O69262 Alpha-L-arabinofuranosidase (EC 3.2.1.55).
Length = 496
Score = 34.7 bits (78), Expect = 0.70
Identities = 35/131 (26%), Positives = 57/131 (43%), Gaps = 4/131 (3%)
Frame = +1
Query: 10 PNYWMLHFFKESSGAILHPTAIQVSSYD---QLVASAITWQNAKDKSTYLRIKVVNFGNQ 180
P Y + + FK A L T V +L +++ A D ++ + ++F
Sbjct: 367 PTYHVFNMFKVHQDAELLDTWESVERTGPEGELPKVSVSASRAADGKIHISLCNLDFETG 426
Query: 181 AVDLNISVAELATGVKKSGSKQTVLTSSSPLDENSFQQPEKVAPVS-SPMANAGQQMGAS 357
A ++I + L GV +G T LTS N+F +PE+V P G + AS
Sbjct: 427 A-SVDIELRGLNGGVSATG---TTLTSGRIDGHNTFDEPERVKPAPFRDFKLEGGHLNAS 482
Query: 358 VGPYSLTSFDL 390
+ P S+T +L
Sbjct: 483 LPPMSVTVLEL 493
>sptr|Q94CQ9|Q94CQ9 Putative beta 1,3-glucanase.
Length = 343
Score = 33.5 bits (75), Expect = 1.6
Identities = 30/110 (27%), Positives = 47/110 (42%), Gaps = 18/110 (16%)
Frame = +1
Query: 94 QLVASAITWQNAKDKSTY--LRIKVVNFGN---------------QAVDLNISVAELATG 222
QL A+A +W K+ Y ++IK + GN Q V ++ A LA
Sbjct: 65 QLAANADSWVQDNVKAYYPDVKIKYIVVGNELTGTGDAASILPAMQNVQAALASAGLADS 124
Query: 223 VKKSGS-KQTVLTSSSPLDENSFQQPEKVAPVSSPMANAGQQMGASVGPY 369
+K + + K L +SSP F P + P+ + G + A+V PY
Sbjct: 125 IKVTTAIKMDTLAASSPPSAGVFTNPSVMEPIVRFLTGNGAPLLANVYPY 174
>sw|Q59219|ABF2_BACOV Alpha-L-arabinofuranosidase 2 (EC 3.2.1.55)
(Arabinosidase 2).
Length = 514
Score = 33.5 bits (75), Expect = 1.6
Identities = 27/105 (25%), Positives = 51/105 (48%), Gaps = 4/105 (3%)
Frame = +1
Query: 10 PNYWMLHFFKESSGAILHP---TAIQVSSYDQLVASAITWQNAKDKSTYLRIKVVNF-GN 177
P Y++ +K A P T ++S D ++ +K+K + I + N +
Sbjct: 386 PTYYVFKMYKVHQDATYLPIDLTCEKMSVRDNRTVPMVSATASKNKDGVIHISLSNVDAD 445
Query: 178 QAVDLNISVAELATGVKKSGSKQTVLTSSSPLDENSFQQPEKVAP 312
+A ++ I++ + T KK+ + +LT+S D NSF++P V P
Sbjct: 446 EAQEITINLGD--TKAKKAIGE--ILTASKLTDYNSFEKPNIVKP 486
>sptr|Q80V54|Q80V54 1300015B04Rik protein.
Length = 608
Score = 33.1 bits (74), Expect = 2.0
Identities = 17/36 (47%), Positives = 23/36 (63%)
Frame = +2
Query: 17 IGCYTFSRNLAAPSSILQPFRSPATISWLRLPSLGR 124
+ C F + + APS+ILQP + PA +S L LP GR
Sbjct: 259 VSCEAFPKAVLAPSAILQPRQHPAKMS-LLLPEAGR 293
>sptr|Q8KN01|Q8KN01 Hypothetical protein Ste20.
Length = 424
Score = 33.1 bits (74), Expect = 2.0
Identities = 22/72 (30%), Positives = 32/72 (44%), Gaps = 3/72 (4%)
Frame = -3
Query: 407 LLGSSSRSNEVRE---YGPTEAPICCPAFAIGLDTGATFSGCWKEFSSSGLLDVRTVCFD 237
+L +S R + +RE YG E C P F +G+ G +G + T D
Sbjct: 268 VLETSGRPHSIREFTPYGYDERQFCSPGFDLGV-------GSLSRTPYAGYPEYHTSADD 320
Query: 236 PDFLTPVASSAT 201
PDF++P A T
Sbjct: 321 PDFVSPEAMEDT 332
>sptr|Q9FKK4|Q9FKK4 Genomic DNA, chromosome 5, P1 clone:MUF9
(AT5g60420/muf9_70).
Length = 873
Score = 32.7 bits (73), Expect = 2.6
Identities = 31/135 (22%), Positives = 56/135 (41%), Gaps = 1/135 (0%)
Frame = +1
Query: 19 WMLHFFKESSGAILHPTAIQVSSYDQLVASAITWQNAKDKSTYLRIKVVNFGNQAVDLNI 198
W + F +ES L + + L SA+ + K K L +K + + L +
Sbjct: 445 WRVKFKRESERRELGELSQWHAPDGSLCPSAV---DIKRKMEMLPVKQEGYSDGPAPLKL 501
Query: 199 SVAELATGVKKSGSKQTVLTSSSPLDENSFQQPEKVAPVSSPMANAGQQ-MGASVGPYSL 375
+ + G+ + T SSS E Q + + P+SS +G+ ASV ++
Sbjct: 502 GIRKNRNGIWEVSKPNTNGLSSSNRQEKVGYQEKNIIPMSSSATGSGRDGDDASVNQDAI 561
Query: 376 TSFDLLLEPSKHDSV 420
+FD + + DS+
Sbjct: 562 GTFDFVANGMELDSI 576
>sptr|O15884|O15884 Invariant surface glycoprotein 100.
Length = 1350
Score = 32.7 bits (73), Expect = 2.6
Identities = 37/133 (27%), Positives = 59/133 (44%), Gaps = 2/133 (1%)
Frame = -3
Query: 503 IIQSNMVSSIFDLSVRTLLSCFFS*QFYTL--SCLLGSSSRSNEVREYGPTEAPICCPAF 330
++ S SS ++ +LLS F S + SCLL S + S+ V + +
Sbjct: 1219 LLSSFASSSAVGIAASSLLSSFASSSAVGIAASCLLSSFASSSAVGMATSSLLSSFASSS 1278
Query: 329 AIGLDTGATFSGCWKEFSSSGLLDVRTVCFDPDFLTPVASSATDMFRSTAWLPKFTTFIL 150
A+G+ T + S F+SS + + T F +SSA M S+ L F +
Sbjct: 1279 AVGMATSSLLSS----FASSSAVGMATSSLLSSF---ASSSAVGMAASSLSLLFFFCWSS 1331
Query: 149 R*VLLSLAFCQVM 111
V+LS+ FC V+
Sbjct: 1332 VIVMLSMTFCDVV 1344
>sptr|Q97DN6|Q97DN6 Probable alpha-arabinofuranosidase.
Length = 835
Score = 32.7 bits (73), Expect = 2.6
Identities = 26/90 (28%), Positives = 41/90 (45%), Gaps = 2/90 (2%)
Frame = +1
Query: 136 KSTYLRIKVVNFGNQAVDLNISVAELATGVKKSGSKQTV--LTSSSPLDENSFQQPEKVA 309
K+ L KVVN ++ + I+V G K S V +T SS D NSF +PE V+
Sbjct: 750 KTNELIAKVVNVADEDRKVRINV----NGSKNISSTANVQYITGSSEDDTNSFDKPENVS 805
Query: 310 PVSSPMANAGQQMGASVGPYSLTSFDLLLE 399
+ + N + YS++ + L+
Sbjct: 806 IKTKKLNNISKNFDYDADKYSVSVIRIKLK 835
>sptr|Q9EVS2|Q9EVS2 FtsZ protein (Cell division protein ftsZ)
(Fragment).
Length = 330
Score = 32.3 bits (72), Expect = 3.5
Identities = 14/48 (29%), Positives = 27/48 (56%)
Frame = +1
Query: 175 NQAVDLNISVAELATGVKKSGSKQTVLTSSSPLDENSFQQPEKVAPVS 318
+QA++ + V+ LATG+ + +V + P +E +F+ P P+S
Sbjct: 266 DQAMEGRVRVSVLATGIDSCNNNSSVNQNKXPAEEKNFKWPYNQIPIS 313
>sptr|Q8AAW4|Q8AAW4 Alpha-L-arabinofuranosidase.
Length = 514
Score = 32.3 bits (72), Expect = 3.5
Identities = 25/107 (23%), Positives = 53/107 (49%), Gaps = 4/107 (3%)
Frame = +1
Query: 10 PNYWMLHFFKESSGAILHP---TAIQVSSYDQLVASAITWQNAKDKSTYLRIKVVNF-GN 177
P Y++ +K A P T +++ D ++ +K+K+ + I + N +
Sbjct: 386 PTYYVFKMYKVHQDATYLPLDLTCEKMNVRDNRTVPMVSATASKNKNGVIHISLSNVDAD 445
Query: 178 QAVDLNISVAELATGVKKSGSKQTVLTSSSPLDENSFQQPEKVAPVS 318
A ++ +++ ++ KK+ + +LTS++ D NSF++P V P S
Sbjct: 446 NAQEITVNLPDV--NAKKAIGE--ILTSANLTDYNSFEKPNIVKPAS 488
>sptr|Q8QLB0|Q8QLB0 LEF-9.
Length = 505
Score = 32.3 bits (72), Expect = 3.5
Identities = 16/47 (34%), Positives = 28/47 (59%)
Frame = +1
Query: 319 SPMANAGQQMGASVGPYSLTSFDLLLEPSKHDSV*NCQLKKHESSVR 459
S M+NA Q+ P S T+FD+L +P+ D+V C + + ++ +R
Sbjct: 7 SAMSNATQRTCVEFVPKSSTTFDILFDPADIDNVFFCNIDQFKTFLR 53
>sptr|Q8JM31|Q8JM31 Putative late expression factor LEF-9.
Length = 497
Score = 32.0 bits (71), Expect = 4.5
Identities = 16/45 (35%), Positives = 27/45 (60%)
Frame = +1
Query: 325 MANAGQQMGASVGPYSLTSFDLLLEPSKHDSV*NCQLKKHESSVR 459
MANA Q+ P S T+FD+L +P+ D+V C + + ++ +R
Sbjct: 1 MANATQRTCIEFVPKSSTTFDILFDPADIDNVFFCNIDQFKTFLR 45
>sptr|Q60086|Q60086 Cell division protein ftsZ (Fragment).
Length = 315
Score = 32.0 bits (71), Expect = 4.5
Identities = 15/48 (31%), Positives = 27/48 (56%)
Frame = +1
Query: 175 NQAVDLNISVAELATGVKKSGSKQTVLTSSSPLDENSFQQPEKVAPVS 318
+QA++ + V+ LATG+ + +V + P +E +F+ P PVS
Sbjct: 259 DQAMEGRVRVSVLATGIDSCNNNSSVNQNKIPAEEKNFKWPYNQIPVS 306
>sptr|Q8FEK9|Q8FEK9 Hypothetical protein ygbK.
Length = 420
Score = 32.0 bits (71), Expect = 4.5
Identities = 15/48 (31%), Positives = 24/48 (50%)
Frame = +1
Query: 40 ESSGAILHPTAIQVSSYDQLVASAITWQNAKDKSTYLRIKVVNFGNQA 183
E+SG + I+ ++ + W NA DK L +K NFG++A
Sbjct: 362 ETSGVVTQSLGIKGFHIGPTISPGVPWVNALDKPVSLALKSGNFGDEA 409
>sptr|Q845Y5|Q845Y5 Cell division protein (Fragment).
Length = 334
Score = 31.6 bits (70), Expect = 5.9
Identities = 14/48 (29%), Positives = 27/48 (56%)
Frame = +1
Query: 175 NQAVDLNISVAELATGVKKSGSKQTVLTSSSPLDENSFQQPEKVAPVS 318
+QA++ + V+ LATG+ + +V + P +E +F+ P P+S
Sbjct: 265 DQAMEGRVRVSVLATGIDSCNNNSSVNQNKIPAEEKNFKWPYNQIPIS 312
>sptr|Q84BU1|Q84BU1 Cell division protein (Fragment).
Length = 334
Score = 31.6 bits (70), Expect = 5.9
Identities = 14/48 (29%), Positives = 27/48 (56%)
Frame = +1
Query: 175 NQAVDLNISVAELATGVKKSGSKQTVLTSSSPLDENSFQQPEKVAPVS 318
+QA++ + V+ LATG+ + +V + P +E +F+ P P+S
Sbjct: 265 DQAMEGRVRVSVLATGIDSCNNNSSVNQNKIPAEEKNFKWPYNQIPIS 312
>sptr|Q8XTM5|Q8XTM5 Hypothetical protein RSp0081.
Length = 199
Score = 31.6 bits (70), Expect = 5.9
Identities = 23/66 (34%), Positives = 32/66 (48%)
Frame = -2
Query: 237 SRLLDAGGQFRNGYVQIHSLVAEIHHLYPEVSALVFSVLPSDGRRNQLIVAGDLNGCRME 58
+R L GG + + ++ V ++HL ALV SV +DG QLI G +E
Sbjct: 51 ARFLVGGGHLSDEMLAAYTQVDYVNHL-----ALVVSVADADGAGEQLIADGRF---VLE 102
Query: 57 DGAARF 40
DG A F
Sbjct: 103 DGVAEF 108
>sptr|Q60090|Q60090 Cell division protein ftsZ (Fragment).
Length = 315
Score = 31.6 bits (70), Expect = 5.9
Identities = 14/48 (29%), Positives = 27/48 (56%)
Frame = +1
Query: 175 NQAVDLNISVAELATGVKKSGSKQTVLTSSSPLDENSFQQPEKVAPVS 318
+QA++ + V+ LATG+ + +V + P +E +F+ P P+S
Sbjct: 259 DQAMEGRVRVSVLATGIDSCNNNSSVNQNKIPAEEKNFKWPYNQIPIS 306
Database: /db/trembl-ebi/tmp/swall
Posted date: Sep 26, 2003 8:29 PM
Number of letters in database: 406,886,216
Number of sequences in database: 1,272,877
Lambda K H
0.318 0.135 0.401
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 413,210,922
Number of Sequences: 1272877
Number of extensions: 8319233
Number of successful extensions: 27959
Number of sequences better than 10.0: 30
Number of HSP's better than 10.0 without gapping: 27114
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 27932
length of database: 406,886,216
effective HSP length: 115
effective length of database: 260,505,361
effective search space used: 16932848465
frameshift window, decay const: 50, 0.1
T: 12
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)

