BLASTX 2.2.6 [Apr-09-2003]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= 3023663.2.1
(613 letters)
Database: /db/uniprot/tmp/swall
1,395,590 sequences; 442,889,342 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
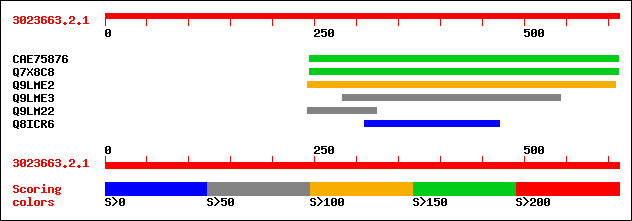
sptrnew|CAE75876|CAE75876 OSJNBa0086B14.27 protein. 179 2e-44
sptr|Q7X8C8|Q7X8C8 OSJNBa0086B14.26 protein (OSJNBa0027G07.1 pro... 179 2e-44
sptr|Q9LME2|Q9LME2 T16E15.12. 105 3e-22
sptr|Q9LME3|Q9LME3 T16E15.11 protein. 64 2e-09
sptr|Q9LM22|Q9LM22 F16L1.1. 54 2e-06
sptr|Q8ICR6|Q8ICR6 Hypothetical protein. 32 8.4
>sptrnew|CAE75876|CAE75876 OSJNBa0086B14.27 protein.
Length = 920
Score = 179 bits (454), Expect = 2e-44
Identities = 92/125 (73%), Positives = 101/125 (80%), Gaps = 2/125 (1%)
Frame = -2
Query: 612 EVQLVSPETNRKDVNLPGILQSPISNMLRKVEKVSQDIPKHRKVTHPEYEVETANGRITK 433
++QLVSPET RKDVNL GI+QSPI+NMLRKVEK +QDIPKHRKVTH EYEVETANGRITK
Sbjct: 795 QLQLVSPETKRKDVNLSGIIQSPITNMLRKVEKGTQDIPKHRKVTHHEYEVETANGRITK 854
Query: 432 RRKTRSPVMFG--EPNSQKPLHNXXXXXXXXXXXVPTRSRAHPANIGELFSEGSLNPYAD 259
RRKT+S VMFG EPN+QK LH+ V S HPANIGELFSEGSLNPYA+
Sbjct: 855 RRKTKSTVMFGVQEPNTQKSLHDTADKDPTKMKKVVAGSHPHPANIGELFSEGSLNPYAE 914
Query: 258 DPYAF 244
DPYAF
Sbjct: 915 DPYAF 919
>sptr|Q7X8C8|Q7X8C8 OSJNBa0086B14.26 protein (OSJNBa0027G07.1
protein).
Length = 894
Score = 179 bits (454), Expect = 2e-44
Identities = 92/125 (73%), Positives = 101/125 (80%), Gaps = 2/125 (1%)
Frame = -2
Query: 612 EVQLVSPETNRKDVNLPGILQSPISNMLRKVEKVSQDIPKHRKVTHPEYEVETANGRITK 433
++QLVSPET RKDVNL GI+QSPI+NMLRKVEK +QDIPKHRKVTH EYEVETANGRITK
Sbjct: 769 QLQLVSPETKRKDVNLSGIIQSPITNMLRKVEKGTQDIPKHRKVTHHEYEVETANGRITK 828
Query: 432 RRKTRSPVMFG--EPNSQKPLHNXXXXXXXXXXXVPTRSRAHPANIGELFSEGSLNPYAD 259
RRKT+S VMFG EPN+QK LH+ V S HPANIGELFSEGSLNPYA+
Sbjct: 829 RRKTKSTVMFGVQEPNTQKSLHDTADKDPTKMKKVVAGSHPHPANIGELFSEGSLNPYAE 888
Query: 258 DPYAF 244
DPYAF
Sbjct: 889 DPYAF 893
>sptr|Q9LME2|Q9LME2 T16E15.12.
Length = 871
Score = 105 bits (263), Expect = 3e-22
Identities = 59/130 (45%), Positives = 75/130 (57%), Gaps = 7/130 (5%)
Frame = -2
Query: 609 VQLVSPETNRKDVNLPGILQSPISNMLRKVEKVSQ----DIPK---HRKVTHPEYEVETA 451
+++ S N +D ++P+S +L+K + V+ IP H KVTH EYEVET
Sbjct: 742 IRVRSDNDNVQDSPFVKAKETPVSKILKKAQNVNAGSVLSIPNPKHHSKVTHREYEVETN 801
Query: 450 NGRITKRRKTRSPVMFGEPNSQKPLHNXXXXXXXXXXXVPTRSRAHPANIGELFSEGSLN 271
NGR+TKRRKTR+ MF EP ++ S A ANIG+LFSEGSLN
Sbjct: 802 NGRVTKRRKTRNTTMFEEPQRRRTRATPKLTPQSIAKGTGMTSHARSANIGDLFSEGSLN 861
Query: 270 PYADDPYAFD 241
PYADDPYAFD
Sbjct: 862 PYADDPYAFD 871
>sptr|Q9LME3|Q9LME3 T16E15.11 protein.
Length = 1025
Score = 63.5 bits (153), Expect = 2e-09
Identities = 40/89 (44%), Positives = 46/89 (51%), Gaps = 2/89 (2%)
Frame = -2
Query: 543 ISNMLRKVEKVSQDIPKHR-KVTHPEYEVETANGRITKRRKTRSPVMFGEPNSQKP-LHN 370
+SN+L++ PKH KVTH EYEVET NGRI KRRKTR MF EP + L
Sbjct: 904 VSNILKEATN-----PKHHSKVTHREYEVETNNGRIPKRRKTRQTTMFQEPQRRSTRLTP 958
Query: 369 XXXXXXXXXXXVPTRSRAHPANIGELFSE 283
H ANIG+LFSE
Sbjct: 959 KLMTPTIIAKETAMADHPHSANIGDLFSE 987
>sptr|Q9LM22|Q9LM22 F16L1.1.
Length = 35
Score = 53.9 bits (128), Expect = 2e-06
Identities = 24/28 (85%), Positives = 25/28 (89%)
Frame = -2
Query: 324 SRAHPANIGELFSEGSLNPYADDPYAFD 241
S A ANIG+LFSEGSLNPYADDPYAFD
Sbjct: 8 SHARSANIGDLFSEGSLNPYADDPYAFD 35
>sptr|Q8ICR6|Q8ICR6 Hypothetical protein.
Length = 2017
Score = 31.6 bits (70), Expect = 8.4
Identities = 21/59 (35%), Positives = 32/59 (54%), Gaps = 5/59 (8%)
Frame = -3
Query: 470 NTKLRQQMEESQSAGKLGAL-----SCLVNRTLRNHCIILLTKMLKN*KRFLQDPVHIL 309
N KL + E S+ ++ A+ +C +N+ N CIILL K++K L DP H+L
Sbjct: 337 NEKLLKTAEPSKMNKEIEAMMNKANNCYINQNF-NECIILLEKVIKK-SPCLHDPFHLL 393
Database: /db/uniprot/tmp/swall
Posted date: Mar 5, 2004 7:45 PM
Number of letters in database: 442,889,342
Number of sequences in database: 1,395,590
Lambda K H
0.318 0.135 0.401
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 520,553,661
Number of Sequences: 1395590
Number of extensions: 11265648
Number of successful extensions: 30197
Number of sequences better than 10.0: 6
Number of HSP's better than 10.0 without gapping: 29390
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 30189
length of database: 442,889,342
effective HSP length: 117
effective length of database: 279,605,312
effective search space used: 24046056832
frameshift window, decay const: 50, 0.1
T: 12
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)

