BLASTX 2.2.6 [Apr-09-2003]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= 2750698.2.1
(1126 letters)
Database: /db/uniprot/tmp/swall
1,395,590 sequences; 442,889,342 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
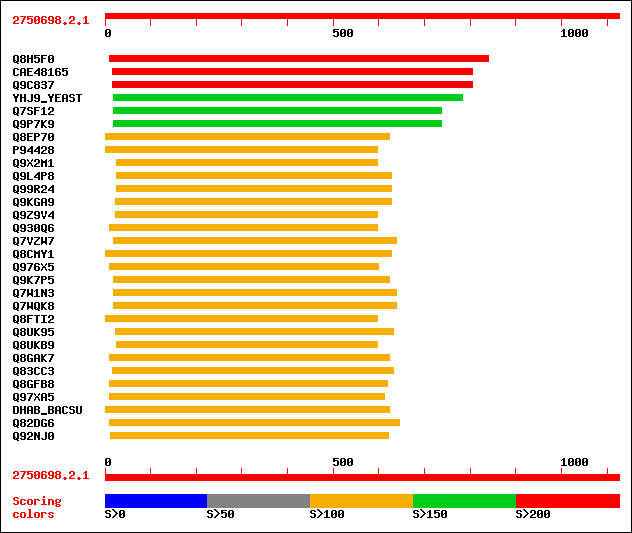
sptr|Q8H5F0|Q8H5F0 OJ1165_F02.19 protein. 499 e-140
sptrnew|CAE48165|CAE48165 Putative aldehyde dehydrogenase (EC 1.... 439 e-122
sptr|Q9C837|Q9C837 Betaine aldehyde dehydrogenase, putative. 398 e-110
sw|P38694|YHJ9_YEAST Hypothetical aldehyde-dehydrogenase like pr... 171 1e-41
sptr|Q7SF12|Q7SF12 Hypothetical protein. 164 2e-39
sptr|Q9P7K9|Q9P7K9 Putative aldehyde-dehydrogenase-like protein. 163 5e-39
sptr|Q8EP70|Q8EP70 Succinate-semialdehyde dehydrogenase (EC 1.2.... 139 1e-31
sptr|P94428|P94428 HOMOLOGUE of succinate semialdehyde dehydroge... 137 4e-31
sptr|Q9X2M1|Q9X2M1 Glycine betaine aldehyde dehydrogenase. 135 1e-30
sptr|Q9L4P8|Q9L4P8 Hypothetical protein (Glycine betaine aldehyd... 134 3e-30
sptr|Q99R24|Q99R24 Glycine betaine aldehyde dehydrogenase gbsA. 134 3e-30
sptr|Q9KGA9|Q9KGA9 Glycine betaine aldehyde dehydrogenase (EC 1.... 133 4e-30
sptr|Q9Z9V4|Q9Z9V4 Aldehyde dehydrogenase. 133 6e-30
sptr|Q930Q6|Q930Q6 GabD3 succinate-semialdehyde dehdyrogenase. 133 6e-30
sptr|Q7VZW7|Q7VZW7 Succinate-semialdehyde dehydrogenase (EC 1.2.... 132 7e-30
sptr|Q8CMY1|Q8CMY1 Glycine betaine aldehyde dehydrogenase gbsA. 132 1e-29
sptr|Q976X5|Q976X5 Putative aldehyde dehydrogenase. 132 1e-29
sptr|Q9K7P5|Q9K7P5 Succinate-semialdehyde dehydrogenase. 132 1e-29
sptr|Q7W1N3|Q7W1N3 Succinate-semialdehyde dehydrogenase (EC 1.2.... 131 2e-29
sptr|Q7WQK8|Q7WQK8 Succinate-semialdehyde dehydrogenase (EC 1.2.... 131 2e-29
sptr|Q8FTI2|Q8FTI2 Putative succinate-semialdehyde dehydrogenase. 131 2e-29
sptr|Q8UK95|Q8UK95 Aldehyde dehydrogenase. 131 2e-29
sptr|Q8UKB9|Q8UKB9 Aldehyde dehydrogenase. 131 2e-29
sptr|Q8GAK7|Q8GAK7 Putative succinate-semialdehyde dehydrogenase. 130 3e-29
sptr|Q83CC3|Q83CC3 Aldehyde dehydrogenase family protein. 130 4e-29
sptr|Q8GFB8|Q8GFB8 Orf17. 130 5e-29
sptr|Q97XA5|Q97XA5 Glyceraldehyde-3-phosphate dehydrogenase, NAD... 129 6e-29
sw|P71016|DHAB_BACSU Betaine aldehyde dehydrogenase (EC 1.2.1.8)... 129 6e-29
sptr|Q82DG6|Q82DG6 Putative aldehyde dehydrogenase. 129 8e-29
sptr|Q92NJ0|Q92NJ0 Putative aldehyde dehydrogenase protein (EC 1... 129 8e-29
>sptr|Q8H5F0|Q8H5F0 OJ1165_F02.19 protein.
Length = 597
Score = 499 bits (1285), Expect = e-140
Identities = 243/277 (87%), Positives = 264/277 (95%)
Frame = +1
Query: 10 AALQSSGQNCAGAERFYVHDDIYSAFVSQIVKTVKSISVGPPLSGRYDMGAICMIEHSEK 189
AALQSSGQNCAGAERFYVH DIYS FVSQ+VK +KSISVGPPLSGRYDMGAICMIEHSEK
Sbjct: 323 AALQSSGQNCAGAERFYVHKDIYSTFVSQVVKIIKSISVGPPLSGRYDMGAICMIEHSEK 382
Query: 190 LQNLVNDALDKGAEIAVRGSFGNLGEDAVDQFFPPTGLVNVDHTMKIMQEETFGPIIPIM 369
LQNLVNDA+DKGAEIA RGSFG+LGEDAVDQFFPPT LVNV+HTMKIMQEE FGPI+PIM
Sbjct: 383 LQNLVNDAVDKGAEIAGRGSFGHLGEDAVDQFFPPTVLVNVNHTMKIMQEEAFGPILPIM 442
Query: 370 KFSSDEEAIKLANDSKYGLGCAVFSGNQKRAIRIASQLHCGVAAINDFASSYMCQSLPFG 549
KF+SDEE +KLANDSKYGLGCAVFSGNQKRAI+IASQLHCGVAAINDFASSYMCQSLPFG
Sbjct: 443 KFNSDEEVVKLANDSKYGLGCAVFSGNQKRAIKIASQLHCGVAAINDFASSYMCQSLPFG 502
Query: 550 GVKDSGFGRFAGVEGLRACCLVKSVVEDRLWPYIRTVIPKPIQYPVSEHGFEFQQLLVET 729
GVKDSGFGRFAGVEGLRACCLVK+VVEDR WPY++T+IPKPIQYPVSE+GFEFQ+LLVET
Sbjct: 503 GVKDSGFGRFAGVEGLRACCLVKAVVEDRWWPYVKTMIPKPIQYPVSENGFEFQELLVET 562
Query: 730 LYGYSVWDRLRSLVNLIKMVTEQNSAPASNATTKKRR 840
LYG SVWDRLRSLVNL+KM++EQN++PA+ T KK R
Sbjct: 563 LYGLSVWDRLRSLVNLLKMISEQNNSPAN--TRKKSR 597
>sptrnew|CAE48165|CAE48165 Putative aldehyde dehydrogenase (EC
1.2.1.3).
Length = 596
Score = 439 bits (1130), Expect = e-122
Identities = 206/263 (78%), Positives = 241/263 (91%)
Frame = +1
Query: 16 LQSSGQNCAGAERFYVHDDIYSAFVSQIVKTVKSISVGPPLSGRYDMGAICMIEHSEKLQ 195
LQSSGQNCAGAERFYVH DIY+AF+ Q+ K VKS+S GPPL+GRYDMGAIC+ EHSE LQ
Sbjct: 325 LQSSGQNCAGAERFYVHKDIYTAFIGQVTKIVKSVSAGPPLTGRYDMGAICLQEHSEHLQ 384
Query: 196 NLVNDALDKGAEIAVRGSFGNLGEDAVDQFFPPTGLVNVDHTMKIMQEETFGPIIPIMKF 375
+LVNDALDKGAEIAVRGSFG+LGEDAVDQ+FPPT L+NV+H MKIM+EE FGPI+PIM+F
Sbjct: 385 SLVNDALDKGAEIAVRGSFGHLGEDAVDQYFPPTVLINVNHNMKIMKEEAFGPIMPIMQF 444
Query: 376 SSDEEAIKLANDSKYGLGCAVFSGNQKRAIRIASQLHCGVAAINDFASSYMCQSLPFGGV 555
S+DEE IKLANDS+Y LGCAVFSG++ RA +IASQ+ CGVAAINDFAS+YMCQSLPFGGV
Sbjct: 445 STDEEVIKLANDSRYALGCAVFSGSKHRAKQIASQIQCGVAAINDFASNYMCQSLPFGGV 504
Query: 556 KDSGFGRFAGVEGLRACCLVKSVVEDRLWPYIRTVIPKPIQYPVSEHGFEFQQLLVETLY 735
KDSGFGRFAG+EGLRACCLVKSVVEDR WP I+T IPKPIQYPV+E+ FEFQ+ LVETLY
Sbjct: 505 KDSGFGRFAGIEGLRACCLVKSVVEDRFWPLIKTKIPKPIQYPVAENAFEFQEALVETLY 564
Query: 736 GYSVWDRLRSLVNLIKMVTEQNS 804
G ++WDRLRSL++++K +T+Q+S
Sbjct: 565 GLNIWDRLRSLIDVLKFLTDQSS 587
>sptr|Q9C837|Q9C837 Betaine aldehyde dehydrogenase, putative.
Length = 578
Score = 398 bits (1023), Expect = e-110
Identities = 192/263 (73%), Positives = 226/263 (85%)
Frame = +1
Query: 16 LQSSGQNCAGAERFYVHDDIYSAFVSQIVKTVKSISVGPPLSGRYDMGAICMIEHSEKLQ 195
LQSSGQNCAGAERFYVH DIY+AF+ Q+ K VKS+S GPPL+GRYDMGAIC+ EHSE LQ
Sbjct: 325 LQSSGQNCAGAERFYVHKDIYTAFIGQVTKIVKSVSAGPPLTGRYDMGAICLQEHSEHLQ 384
Query: 196 NLVNDALDKGAEIAVRGSFGNLGEDAVDQFFPPTGLVNVDHTMKIMQEETFGPIIPIMKF 375
+LVNDALDKGAEIAVRGSFG+LGEDAVDQ+FPPT L+NV+H MKIM+EE FGPI+PIM+F
Sbjct: 385 SLVNDALDKGAEIAVRGSFGHLGEDAVDQYFPPTVLINVNHNMKIMKEEAFGPIMPIMQF 444
Query: 376 SSDEEAIKLANDSKYGLGCAVFSGNQKRAIRIASQLHCGVAAINDFASSYMCQSLPFGGV 555
S+DEE IKLANDS+Y LGCAVFSG++ RA +IASQ+ CGVAAINDFAS+YMCQSLPFGGV
Sbjct: 445 STDEEVIKLANDSRYALGCAVFSGSKHRAKQIASQIQCGVAAINDFASNYMCQSLPFGGV 504
Query: 556 KDSGFGRFAGVEGLRACCLVKSVVEDRLWPYIRTVIPKPIQYPVSEHGFEFQQLLVETLY 735
KDSGFGRFAG+EGLRACCLVKSV YPV+E+ FEFQ+ LVETLY
Sbjct: 505 KDSGFGRFAGIEGLRACCLVKSV------------------YPVAENAFEFQEALVETLY 546
Query: 736 GYSVWDRLRSLVNLIKMVTEQNS 804
G ++WDRLRSL++++K +T+Q+S
Sbjct: 547 GLNIWDRLRSLIDVLKFLTDQSS 569
>sw|P38694|YHJ9_YEAST Hypothetical aldehyde-dehydrogenase like protein
in FIL1-VMA10 intergenic region (EC 1.2.1.-).
Length = 644
Score = 171 bits (434), Expect = 1e-41
Identities = 104/265 (39%), Positives = 144/265 (54%), Gaps = 10/265 (3%)
Frame = +1
Query: 19 QSSGQNCAGAERFYVHDDIYSAFVSQIVKTVKSISVGPPLSG--------RYDMGAICMI 174
QSSGQNC G ER V + Y +VK + PL DMGA+
Sbjct: 383 QSSGQNCIGIERVIVSKENYD----DLVKILNDRMTANPLRQGSDIDHLENVDMGAMISD 438
Query: 175 EHSEKLQNLVNDALDKGAEIAVRGSFGNLGEDAVDQFFPPTGLVNVDHTMKIMQEETFGP 354
++L+ LV DA+ KGA + GS + +F PT LV+V MKI Q E FGP
Sbjct: 439 NRFDELEALVKDAVAKGARLLQGGSRFKHPKYPQGHYFQPTLLVDVTPEMKIAQNEVFGP 498
Query: 355 IIPIMKFSSDEEAIKLANDSKYGLGCAVFSGNQKRAIRIASQLHCGVAAINDFASSYMCQ 534
I+ +MK + + ++LAN + +GLG +VF + K +A+ L G AINDFA+ Y+CQ
Sbjct: 499 ILVMMKAKNTDHCVQLANSAPFGLGGSVFGADIKECNYVANSLQTGNVAINDFATFYVCQ 558
Query: 535 SLPFGGVKDSGFGRFAGVEGLRACCLVKSVVEDRLWPYIRTVIPKPIQYPVSEH--GFEF 708
LPFGG+ SG+G+F G EGL C KSV D L P++ T IPKP+ YP+ + + F
Sbjct: 559 -LPFGGINGSGYGKFGGEEGLLGLCNAKSVCFDTL-PFVSTQIPKPLDYPIRNNAKAWNF 616
Query: 709 QQLLVETLYGYSVWDRLRSLVNLIK 783
+ + Y S W R++SL +L K
Sbjct: 617 VKSFIVGAYTNSTWQRIKSLFSLAK 641
>sptr|Q7SF12|Q7SF12 Hypothetical protein.
Length = 616
Score = 164 bits (415), Expect = 2e-39
Identities = 100/246 (40%), Positives = 139/246 (56%), Gaps = 6/246 (2%)
Frame = +1
Query: 19 QSSGQNCAGAERFYVHDDIYSAFVSQIVKTVKSISVGPPLSGRYDMGAICMIEHSEKLQN 198
Q+SGQNC G ER V Y +S + V+++ +GP D+GA+ +L+
Sbjct: 349 QASGQNCIGIERIVVAPQHYDTLLSMLTPRVRALRLGPTA----DVGAMISDNAFARLEG 404
Query: 199 LVNDALDKGAEIAVRGSFGNLGEDAVDQFFPPTGLVNVDHTMKIMQEETFGPIIPIMKF- 375
LV DA+ +GA + G E +F PT LV+V M I QEE FGPI+ +M+
Sbjct: 405 LVADAVKQGARLLAGGKRYAHPEYPSGHYFVPTLLVDVTPDMAIAQEECFGPIMVVMRAA 464
Query: 376 -SSDEEAIKLANDSKYGLGCAVFSGNQKRAIR-IASQLHCGVAAINDFASSYMCQSLPFG 549
SS E+ + +AN +GLG +VF + + L G+ A+NDF ++Y Q LPFG
Sbjct: 465 SSSAEDILAVANAPDFGLGSSVFGSEWDSTLHEVVRGLKAGMVAVNDFGATYAVQ-LPFG 523
Query: 550 GVKDSGFGRFAGVEGLRACCLVKSVVEDRL-WPYIRTVIPKPIQYPV--SEHGFEFQQLL 720
GV SG+GRFAG EGLR C +K+V EDR W +RT IP+P+QYPV E + F + +
Sbjct: 524 GVAGSGYGRFAGEEGLRGLCNIKAVCEDRFGWLGVRTAIPRPMQYPVPDQERSWRFARGV 583
Query: 721 VETLYG 738
VE YG
Sbjct: 584 VEVGYG 589
>sptr|Q9P7K9|Q9P7K9 Putative aldehyde-dehydrogenase-like protein.
Length = 522
Score = 163 bits (412), Expect = 5e-39
Identities = 88/242 (36%), Positives = 138/242 (57%), Gaps = 2/242 (0%)
Frame = +1
Query: 19 QSSGQNCAGAERFYVHDDIYSAFVSQIVKTVKSISVGPPLSGRYDMGAICMIEHSEKLQN 198
QS+GQNC G ER D +Y ++++ + ++ +G DMGA+ + L++
Sbjct: 267 QSAGQNCIGIERIIALDGVYDTIITKLYNRISTMRLGMYTQNDVDMGAMVSNNRFDHLES 326
Query: 199 LVNDALDKGAEIAVRGSFGNLGEDAVDQFFPPTGLVNVDHTMKIMQEETFGPIIPIMKFS 378
L+ DA+ KGA + G + +F PT LV+ + MKI QEE F PI + +
Sbjct: 327 LIQDAVSKGARLVYGGHRFQHPKYPKGNYFLPTLLVDATNEMKIAQEECFAPIALVFRAK 386
Query: 379 SDEEAIKLANDSKYGLGCAVFSGNQKRAIRIASQLHCGVAAINDFASSYMCQSLPFGGVK 558
S E A+++AN +++GLG +VF +++ L G+ A+NDF + Y+ Q +PFGG K
Sbjct: 387 SPEHALEIANGTEFGLGASVFGRDKQLCQYFTDNLETGMVAVNDFGAFYLLQ-MPFGGCK 445
Query: 559 DSGFGRFAGVEGLRACCLVKSVVEDRLWPYIRTVIPKPIQYPV--SEHGFEFQQLLVETL 732
SG+GRFAG EGLR C K++ DR + I T IP + YP+ S+ ++F + L+ T+
Sbjct: 446 KSGYGRFAGYEGLRGICNSKAIAYDR-FSAIHTGIPPAVDYPIPDSQKAWQFVRGLMGTV 504
Query: 733 YG 738
YG
Sbjct: 505 YG 506
>sptr|Q8EP70|Q8EP70 Succinate-semialdehyde dehydrogenase (EC
1.2.1.16).
Length = 462
Score = 139 bits (349), Expect = 1e-31
Identities = 80/208 (38%), Positives = 112/208 (53%)
Frame = +1
Query: 1 ARGAALQSSGQNCAGAERFYVHDDIYSAFVSQIVKTVKSISVGPPLSGRYDMGAICMIEH 180
A + +++GQ C R YVH+ I F+ + V+ + VG L D+G + +
Sbjct: 257 ATASKFRNAGQTCVCTNRIYVHESIKDEFIKAFERKVRELRVGNGLEEGIDIGPLIDHDA 316
Query: 181 SEKLQNLVNDALDKGAEIAVRGSFGNLGEDAVDQFFPPTGLVNVDHTMKIMQEETFGPII 360
EK++ + DA KGA I G +G+D++ F+ PT L NV M M EETFGP+
Sbjct: 317 LEKVETHIRDAAHKGASIVYGGD--KVGDDSL--FYKPTILSNVTDNMLCMTEETFGPVA 372
Query: 361 PIMKFSSDEEAIKLANDSKYGLGCAVFSGNQKRAIRIASQLHCGVAAINDFASSYMCQSL 540
P+ FS++EEA+K AN S YGL +F+ N AIRI+ QL G+ IND S
Sbjct: 373 PVTTFSTEEEAVKRANHSPYGLAAYLFTENLATAIRISEQLEYGIVGINDGVPS--TAQA 430
Query: 541 PFGGVKDSGFGRFAGVEGLRACCLVKSV 624
PFGG K+SG GR G G+ VK +
Sbjct: 431 PFGGFKESGIGREGGHHGMEEYVEVKYI 458
>sptr|P94428|P94428 HOMOLOGUE of succinate semialdehyde
dehydrogenase GABD of E. COLI.
Length = 462
Score = 137 bits (344), Expect = 4e-31
Identities = 76/199 (38%), Positives = 114/199 (57%)
Frame = +1
Query: 1 ARGAALQSSGQNCAGAERFYVHDDIYSAFVSQIVKTVKSISVGPPLSGRYDMGAICMIEH 180
A + +++GQ C A R VH+ I F +++ + V + VG L ++G I
Sbjct: 254 AMASKYRNAGQTCVCANRLIVHESIKDEFAAKLSEQVSKLKVGNGLEEGVNVGPIINKRG 313
Query: 181 SEKLQNLVNDALDKGAEIAVRGSFGNLGEDAVDQFFPPTGLVNVDHTMKIMQEETFGPII 360
EK+ + ++DA++KGA++ G++ +D F PT L +VD +M IM EETFGP+
Sbjct: 314 FEKIVSQIDDAVEKGAKVIAGGTYDR-NDDKGCYFVNPTVLTDVDTSMNIMHEETFGPVA 372
Query: 361 PIMKFSSDEEAIKLANDSKYGLGCAVFSGNQKRAIRIASQLHCGVAAINDFASSYMCQSL 540
PI+ FS +EAI+LAND+ YGL F+ N +R I I+ L G+ ND S +
Sbjct: 373 PIVTFSDIDEAIQLANDTPYGLAAYFFTENYRRGIYISENLEYGIIGWNDGGPSAV--QA 430
Query: 541 PFGGVKDSGFGRFAGVEGL 597
PFGG+K+SG GR G EG+
Sbjct: 431 PFGGMKESGIGREGGSEGI 449
>sptr|Q9X2M1|Q9X2M1 Glycine betaine aldehyde dehydrogenase.
Length = 497
Score = 135 bits (340), Expect = 1e-30
Identities = 76/191 (39%), Positives = 111/191 (58%)
Frame = +1
Query: 25 SGQNCAGAERFYVHDDIYSAFVSQIVKTVKSISVGPPLSGRYDMGAICMIEHSEKLQNLV 204
+GQ C+ R VH+DI F + +++ +K+I +G +MG + EH EK++N +
Sbjct: 285 AGQVCSAGARIIVHNDIKEKFEAALIERIKNIKLGNGFDEDTEMGPVISAEHREKIENYM 344
Query: 205 NDALDKGAEIAVRGSFGNLGEDAVDQFFPPTGLVNVDHTMKIMQEETFGPIIPIMKFSSD 384
A + A IA+ G + FF PT + + D +M+I+QEE FGP++ I F+++
Sbjct: 345 EVAKSENATIAIGGKRPEREDLQDGFFFEPTVITDCDTSMRIVQEEVFGPVVTIESFTTE 404
Query: 385 EEAIKLANDSKYGLGCAVFSGNQKRAIRIASQLHCGVAAINDFASSYMCQSLPFGGVKDS 564
EAIKLANDS YGL VF+ + +A RIA++L G INDF Y Q+ P+GG K S
Sbjct: 405 AEAIKLANDSIYGLAGGVFTNDVGKAQRIANKLKMGTVWINDF-HPYFAQA-PWGGYKQS 462
Query: 565 GFGRFAGVEGL 597
G GR G GL
Sbjct: 463 GIGRELGKAGL 473
>sptr|Q9L4P8|Q9L4P8 Hypothetical protein (Glycine betaine aldehyde
dehydrogenase gbsA).
Length = 496
Score = 134 bits (337), Expect = 3e-30
Identities = 77/201 (38%), Positives = 114/201 (56%)
Frame = +1
Query: 25 SGQNCAGAERFYVHDDIYSAFVSQIVKTVKSISVGPPLSGRYDMGAICMIEHSEKLQNLV 204
+GQ C+ R V + I F ++ VK I +G +MG + EH K+++ +
Sbjct: 285 AGQVCSAGSRILVQNSIKDKFEQALIDRVKKIKLGNGFDADTEMGPVISTEHRNKIESYM 344
Query: 205 NDALDKGAEIAVRGSFGNLGEDAVDQFFPPTGLVNVDHTMKIMQEETFGPIIPIMKFSSD 384
+ A +GA IAV G + + FF PT + N D +M+I+QEE FGP++ + F ++
Sbjct: 345 DVAKAEGATIAVGGKRPDRDDLKDGLFFEPTVITNCDTSMRIVQEEVFGPVVTVEGFETE 404
Query: 385 EEAIKLANDSKYGLGCAVFSGNQKRAIRIASQLHCGVAAINDFASSYMCQSLPFGGVKDS 564
+EAI+LANDS YGL AVFS + +A R+A++L G INDF Y Q+ P+GG K S
Sbjct: 405 QEAIQLANDSIYGLAGAVFSKDIGKAQRVANKLKLGTVWINDF-HPYFAQA-PWGGYKQS 462
Query: 565 GFGRFAGVEGLRACCLVKSVV 627
G GR G EGL + K ++
Sbjct: 463 GIGRELGKEGLEEYLVSKHIL 483
>sptr|Q99R24|Q99R24 Glycine betaine aldehyde dehydrogenase gbsA.
Length = 496
Score = 134 bits (337), Expect = 3e-30
Identities = 77/201 (38%), Positives = 114/201 (56%)
Frame = +1
Query: 25 SGQNCAGAERFYVHDDIYSAFVSQIVKTVKSISVGPPLSGRYDMGAICMIEHSEKLQNLV 204
+GQ C+ R V + I F ++ VK I +G +MG + EH K+++ +
Sbjct: 285 AGQVCSAGSRILVQNSIKDKFEQALIDRVKKIKLGNGFDADTEMGPVISTEHRNKIESYM 344
Query: 205 NDALDKGAEIAVRGSFGNLGEDAVDQFFPPTGLVNVDHTMKIMQEETFGPIIPIMKFSSD 384
+ A +GA IAV G + + FF PT + N D +M+I+QEE FGP++ + F ++
Sbjct: 345 DVAKAEGATIAVGGKRPDRDDLKDGLFFEPTVITNCDTSMRIVQEEVFGPVVTVEGFETE 404
Query: 385 EEAIKLANDSKYGLGCAVFSGNQKRAIRIASQLHCGVAAINDFASSYMCQSLPFGGVKDS 564
+EAI+LANDS YGL AVFS + +A R+A++L G INDF Y Q+ P+GG K S
Sbjct: 405 QEAIQLANDSIYGLAGAVFSKDIGKAQRVANKLKLGTVWINDF-HPYFAQA-PWGGYKQS 462
Query: 565 GFGRFAGVEGLRACCLVKSVV 627
G GR G EGL + K ++
Sbjct: 463 GIGRELGKEGLEEYLVSKHIL 483
>sptr|Q9KGA9|Q9KGA9 Glycine betaine aldehyde dehydrogenase (EC
1.2.1.8).
Length = 490
Score = 133 bits (335), Expect = 4e-30
Identities = 71/202 (35%), Positives = 113/202 (55%)
Frame = +1
Query: 22 SSGQNCAGAERFYVHDDIYSAFVSQIVKTVKSISVGPPLSGRYDMGAICMIEHSEKLQNL 201
++GQ+C R +VH++IY F+ + + K + +G P +G+I + E + +
Sbjct: 278 NTGQSCEARSRLFVHEEIYDEFIEEFITRAKKLQLGDPFDKGTHVGSIISRDQLEIIDSY 337
Query: 202 VNDALDKGAEIAVRGSFGNLGEDAVDQFFPPTGLVNVDHTMKIMQEETFGPIIPIMKFSS 381
V A ++GA IA+ G + ++ PT + NV MK +QEE FGP++ + F
Sbjct: 338 VRSAEEEGATIALGGKERRVEGFENGHWYEPTVITNVTPDMKAVQEEIFGPVVVVETFQD 397
Query: 382 DEEAIKLANDSKYGLGCAVFSGNQKRAIRIASQLHCGVAAINDFASSYMCQSLPFGGVKD 561
+ E IK AND+K+GLG A+++ +Q RA R+A QL G+ +N S++ PFGG K+
Sbjct: 398 EREVIKRANDTKFGLGSAIWTKDQGRATRVAHQLEAGIVMVNCPFSAF--PGTPFGGYKE 455
Query: 562 SGFGRFAGVEGLRACCLVKSVV 627
SGFGR VE L KS++
Sbjct: 456 SGFGRELCVETLDLYMEDKSIL 477
>sptr|Q9Z9V4|Q9Z9V4 Aldehyde dehydrogenase.
Length = 488
Score = 133 bits (334), Expect = 6e-30
Identities = 69/192 (35%), Positives = 109/192 (56%)
Frame = +1
Query: 22 SSGQNCAGAERFYVHDDIYSAFVSQIVKTVKSISVGPPLSGRYDMGAICMIEHSEKLQNL 201
++GQ+C R +VH++IY F+ + + K + +G P +G+I + E + +
Sbjct: 293 NTGQSCEARSRLFVHEEIYDEFIEEFITRAKKLQLGDPFDKGTHVGSIISRDQLEIIDSY 352
Query: 202 VNDALDKGAEIAVRGSFGNLGEDAVDQFFPPTGLVNVDHTMKIMQEETFGPIIPIMKFSS 381
V A ++GA IA+ G + ++ PT + NV MK +QEE FGP++ + F
Sbjct: 353 VRSAEEEGATIALGGKERRVEGFENGHWYEPTVITNVTPDMKAVQEEIFGPVVVVETFQD 412
Query: 382 DEEAIKLANDSKYGLGCAVFSGNQKRAIRIASQLHCGVAAINDFASSYMCQSLPFGGVKD 561
+ E IK AND+K+GLG A+++ +Q RA R+A QL G+ +N S++ PFGG K+
Sbjct: 413 EREVIKRANDTKFGLGSAIWTKDQGRATRVAHQLEAGIVMVNCPFSAF--PGTPFGGYKE 470
Query: 562 SGFGRFAGVEGL 597
SGFGR VE L
Sbjct: 471 SGFGRELCVETL 482
>sptr|Q930Q6|Q930Q6 GabD3 succinate-semialdehyde dehdyrogenase.
Length = 487
Score = 133 bits (334), Expect = 6e-30
Identities = 75/196 (38%), Positives = 111/196 (56%)
Frame = +1
Query: 10 AALQSSGQNCAGAERFYVHDDIYSAFVSQIVKTVKSISVGPPLSGRYDMGAICMIEHSEK 189
A ++++G+ C A RFYV I+ AFV+ + +KS+ +GP G + K
Sbjct: 285 AKMRNAGEACTAANRFYVQAGIHDAFVAGLTARMKSLKLGPGYDPETQCGPMITQNAVRK 344
Query: 190 LQNLVNDALDKGAEIAVRGSFGNLGEDAVDQFFPPTGLVNVDHTMKIMQEETFGPIIPIM 369
+ LV++AL GA G L E+ F+PPT L NV I +EE FGP+ P+
Sbjct: 345 IDRLVSEALAAGARATTGGK--PLTENGY--FYPPTVLENVPVNASIAREEIFGPVAPVY 400
Query: 370 KFSSDEEAIKLANDSKYGLGCAVFSGNQKRAIRIASQLHCGVAAINDFASSYMCQSLPFG 549
KF SD+EAI+LAN+++YGL ++S + KRA+++ ++ G+ IN S + PFG
Sbjct: 401 KFESDDEAIRLANNTEYGLAAYIYSRDLKRAMKVGKRIETGMLGINRGLMS--DPAAPFG 458
Query: 550 GVKDSGFGRFAGVEGL 597
GVK SG GR GV G+
Sbjct: 459 GVKQSGLGREGGVTGI 474
>sptr|Q7VZW7|Q7VZW7 Succinate-semialdehyde dehydrogenase (EC
1.2.1.16).
Length = 493
Score = 132 bits (333), Expect = 7e-30
Identities = 72/207 (34%), Positives = 113/207 (54%)
Frame = +1
Query: 19 QSSGQNCAGAERFYVHDDIYSAFVSQIVKTVKSISVGPPLSGRYDMGAICMIEHSEKLQN 198
++ GQ C R +V ++ AFV ++ V+ + VGP +G + EK+++
Sbjct: 288 RNGGQTCVCPNRIFVQAGVHDAFVRKLAARVEGLVVGPASDPASQIGPMINARAVEKIEH 347
Query: 199 LVNDALDKGAEIAVRGSFGNLGEDAVDQFFPPTGLVNVDHTMKIMQEETFGPIIPIMKFS 378
V DAL++GA +AV G + A D ++ PT L VD TM +EETFGP+ PI F+
Sbjct: 348 HVADALERGARLAVGGKRIHSARCAGDHYYAPTVLTGVDETMACFREETFGPVAPITVFA 407
Query: 379 SDEEAIKLANDSKYGLGCAVFSGNQKRAIRIASQLHCGVAAINDFASSYMCQSLPFGGVK 558
S+ E + AN + +GL +S + +R R+A L G+ +N+ A + ++ PFGGVK
Sbjct: 408 SEAEVVAAANATAFGLAAYFYSTDVRRIWRLADALETGIVGVNEGALA--AEAAPFGGVK 465
Query: 559 DSGFGRFAGVEGLRACCLVKSVVEDRL 639
DSG+GR GL +K V + +L
Sbjct: 466 DSGYGREGSRHGLAEYMQIKYVCQGQL 492
>sptr|Q8CMY1|Q8CMY1 Glycine betaine aldehyde dehydrogenase gbsA.
Length = 496
Score = 132 bits (332), Expect = 1e-29
Identities = 76/209 (36%), Positives = 113/209 (54%)
Frame = +1
Query: 1 ARGAALQSSGQNCAGAERFYVHDDIYSAFVSQIVKTVKSISVGPPLSGRYDMGAICMIEH 180
A A +GQ C+ R VH+DI F ++ V I +G +MG + H
Sbjct: 277 ALNAGYFHAGQVCSAGSRILVHNDIKDKFEKALIDRVSKIKLGNGFDQDTEMGPVISTAH 336
Query: 181 SEKLQNLVNDALDKGAEIAVRGSFGNLGEDAVDQFFPPTGLVNVDHTMKIMQEETFGPII 360
+K++ + A GA IA+ G + FF PT + + D +M+I+QEE FGP++
Sbjct: 337 RDKIEGYMEVAKKDGATIAIGGKRPEREDLQAGLFFEPTVITDCDTSMRIVQEEVFGPVV 396
Query: 361 PIMKFSSDEEAIKLANDSKYGLGCAVFSGNQKRAIRIASQLHCGVAAINDFASSYMCQSL 540
+ F+ +EEAI+LANDS YGL A+F+ + +A R+A++L G INDF Y Q+
Sbjct: 397 TVEGFADEEEAIRLANDSIYGLAGAIFTKDIGKAQRVANKLKLGTVWINDF-HPYFAQA- 454
Query: 541 PFGGVKDSGFGRFAGVEGLRACCLVKSVV 627
P+GG K SG GR G EGL + K ++
Sbjct: 455 PWGGYKQSGVGRELGKEGLEEYLVSKHIL 483
>sptr|Q976X5|Q976X5 Putative aldehyde dehydrogenase.
Length = 468
Score = 132 bits (331), Expect = 1e-29
Identities = 77/197 (39%), Positives = 107/197 (54%)
Frame = +1
Query: 10 AALQSSGQNCAGAERFYVHDDIYSAFVSQIVKTVKSISVGPPLSGRYDMGAICMIEHSEK 189
A + +GQNC +R V ++IY FV + VK++ VG PL D+G + E EK
Sbjct: 265 ARYEYAGQNCNAGKRIIVREEIYDKFVKAFKEKVKALKVGDPLDESTDIGPVINQESVEK 324
Query: 190 LQNLVNDALDKGAEIAVRGSFGNLGEDAVDQFFPPTGLVNVDHTMKIMQEETFGPIIPIM 369
L + DA KG + V G FFP + + N M +++ E FGPI PI+
Sbjct: 325 LNKALEDAQSKGGNVEVLNKGPETG-----YFFPLSLVTNPSLDMLVLKTEIFGPIAPIV 379
Query: 370 KFSSDEEAIKLANDSKYGLGCAVFSGNQKRAIRIASQLHCGVAAINDFASSYMCQSLPFG 549
SDEEAI +AN ++YGL A+FS + RA++IA +L G IND ++ SLPFG
Sbjct: 380 SVKSDEEAINIANSTEYGLQSAIFSNDVNRALKIAKELKFGAIIIND-STRLRWDSLPFG 438
Query: 550 GVKDSGFGRFAGVEGLR 600
G K +G GR EG+R
Sbjct: 439 GFKKTGIGR----EGVR 451
>sptr|Q9K7P5|Q9K7P5 Succinate-semialdehyde dehydrogenase.
Length = 475
Score = 132 bits (331), Expect = 1e-29
Identities = 77/202 (38%), Positives = 111/202 (54%)
Frame = +1
Query: 19 QSSGQNCAGAERFYVHDDIYSAFVSQIVKTVKSISVGPPLSGRYDMGAICMIEHSEKLQN 198
+++GQ C R YV + I FV ++ V + VG L +G + + EK++
Sbjct: 275 RNAGQTCVCGNRIYVQETIVDEFVKRLTGKVVQLKVGNGLEEGVHIGPLIEKKGYEKVKA 334
Query: 199 LVNDALDKGAEIAVRGSFGNLGEDAVDQFFPPTGLVNVDHTMKIMQEETFGPIIPIMKFS 378
V+DA+ KGA + + G G G D+ F+ PT L +V M +MQEETFGP+ PI F+
Sbjct: 335 HVDDAVAKGARVVIGGK-GQEGNDSY--FYDPTILTDVHDEMLVMQEETFGPVAPIQTFA 391
Query: 379 SDEEAIKLANDSKYGLGCAVFSGNQKRAIRIASQLHCGVAAINDFASSYMCQSLPFGGVK 558
+DE+ I+ AN ++YGL F+ N R IR++ L G+ ND A S PFGG+K
Sbjct: 392 TDEDVIEKANGTRYGLAAYFFTENYARGIRLSEALDFGIVGWNDGAPS--IAQAPFGGMK 449
Query: 559 DSGFGRFAGVEGLRACCLVKSV 624
+SG GR G EG+ A K V
Sbjct: 450 ESGLGREGGQEGIEAFLETKFV 471
>sptr|Q7W1N3|Q7W1N3 Succinate-semialdehyde dehydrogenase (EC
1.2.1.16).
Length = 493
Score = 131 bits (330), Expect = 2e-29
Identities = 71/207 (34%), Positives = 113/207 (54%)
Frame = +1
Query: 19 QSSGQNCAGAERFYVHDDIYSAFVSQIVKTVKSISVGPPLSGRYDMGAICMIEHSEKLQN 198
++ GQ C R +V ++ AFV ++ V+ + VGP +G + EK+++
Sbjct: 288 RNGGQTCVCPNRIFVQAGVHDAFVRKLAARVEGLVVGPASDPASQIGPMINARAVEKIEH 347
Query: 199 LVNDALDKGAEIAVRGSFGNLGEDAVDQFFPPTGLVNVDHTMKIMQEETFGPIIPIMKFS 378
V DAL++GA +AV G + A D ++ PT L VD TM +EETFGP+ PI F+
Sbjct: 348 HVADALERGARLAVGGKRIHSARCAGDHYYAPTVLTGVDETMACFREETFGPVAPITVFA 407
Query: 379 SDEEAIKLANDSKYGLGCAVFSGNQKRAIRIASQLHCGVAAINDFASSYMCQSLPFGGVK 558
S+ + + AN + +GL +S + +R R+A L G+ +N+ A + ++ PFGGVK
Sbjct: 408 SEADVVAAANATAFGLAAYFYSTDVRRIWRLADALETGIVGVNEGALA--AEAAPFGGVK 465
Query: 559 DSGFGRFAGVEGLRACCLVKSVVEDRL 639
DSG+GR GL +K V + +L
Sbjct: 466 DSGYGREGSRHGLAEYMQIKYVCQGQL 492
>sptr|Q7WQK8|Q7WQK8 Succinate-semialdehyde dehydrogenase (EC
1.2.1.16).
Length = 493
Score = 131 bits (330), Expect = 2e-29
Identities = 71/207 (34%), Positives = 113/207 (54%)
Frame = +1
Query: 19 QSSGQNCAGAERFYVHDDIYSAFVSQIVKTVKSISVGPPLSGRYDMGAICMIEHSEKLQN 198
++ GQ C R +V ++ AFV ++ V+ + VGP +G + EK+++
Sbjct: 288 RNGGQTCVCPNRIFVQAGVHDAFVRKLAARVEGLVVGPASDPASQIGPMINARAVEKIEH 347
Query: 199 LVNDALDKGAEIAVRGSFGNLGEDAVDQFFPPTGLVNVDHTMKIMQEETFGPIIPIMKFS 378
V DAL++GA +AV G + A D ++ PT L VD TM +EETFGP+ PI F+
Sbjct: 348 HVADALERGARLAVGGKRIHSARCAGDHYYAPTVLTGVDETMACFREETFGPVAPITVFA 407
Query: 379 SDEEAIKLANDSKYGLGCAVFSGNQKRAIRIASQLHCGVAAINDFASSYMCQSLPFGGVK 558
S+ + + AN + +GL +S + +R R+A L G+ +N+ A + ++ PFGGVK
Sbjct: 408 SEADVVAAANATAFGLAAYFYSTDVRRIWRLADALETGIVGVNEGALA--AEAAPFGGVK 465
Query: 559 DSGFGRFAGVEGLRACCLVKSVVEDRL 639
DSG+GR GL +K V + +L
Sbjct: 466 DSGYGREGSRHGLAEYMQIKYVCQGQL 492
>sptr|Q8FTI2|Q8FTI2 Putative succinate-semialdehyde dehydrogenase.
Length = 540
Score = 131 bits (329), Expect = 2e-29
Identities = 75/199 (37%), Positives = 113/199 (56%)
Frame = +1
Query: 1 ARGAALQSSGQNCAGAERFYVHDDIYSAFVSQIVKTVKSISVGPPLSGRYDMGAICMIEH 180
A GA +++ G+ C A RF VHD I F ++ + +++G L +G + +
Sbjct: 330 AMGAKMRNIGEACTAANRFLVHDSIAQDFSRRLAERFNQLTLGNGLDDGVTVGPLVEAKA 389
Query: 181 SEKLQNLVNDALDKGAEIAVRGSFGNLGEDAVDQFFPPTGLVNVDHTMKIMQEETFGPII 360
E + LV+DA+ +GA + G+ G GE F+ PT LV+V +I+ EE FGP+
Sbjct: 390 RENVAKLVDDAVTEGATVLTGGTAGE-GEG---YFYQPTVLVDVAPGARILTEEIFGPVA 445
Query: 361 PIMKFSSDEEAIKLANDSKYGLGCAVFSGNQKRAIRIASQLHCGVAAINDFASSYMCQSL 540
PI+ F+++EEAI+LAND++YGL +F+ + R RI+ QL G+ N S +
Sbjct: 446 PIITFTTEEEAIQLANDTEYGLASYLFTRDYARMFRISDQLEFGLLGFNSGVISN--AAA 503
Query: 541 PFGGVKDSGFGRFAGVEGL 597
PFGGVK SG GR G EG+
Sbjct: 504 PFGGVKQSGIGREGGTEGI 522
>sptr|Q8UK95|Q8UK95 Aldehyde dehydrogenase.
Length = 507
Score = 131 bits (329), Expect = 2e-29
Identities = 70/205 (34%), Positives = 114/205 (55%), Gaps = 1/205 (0%)
Frame = +1
Query: 22 SSGQNCAGAERFYVHDDIYSAFVSQIVKTVKSISVGPPLSGRYDMGAICMIEHSEKLQNL 201
++GQ C R +V +IY F+ +V+ ++I +G P+ D+G + + KL+
Sbjct: 292 AAGQTCVAGSRCFVEANIYDRFIEALVERTRNIKLGDPMDDATDIGPLALESQLSKLEGY 351
Query: 202 VNDALDKGAEIAVRGSFGNLGEDAVDQFFPPTGLVNVDHTMKIMQEETFGPIIPIMKFSS 381
V + +G+ IA+ G E + +F PT + + + M M+ E FGP++ +M FSS
Sbjct: 352 VASGVREGSRIAIGGKRPQSEELSRGWYFEPTVMADATNDMSFMRHELFGPMVGVMPFSS 411
Query: 382 DEEAIKLANDSKYGLGCAVFSGNQKRAIRIASQLHCGVAAINDFAS-SYMCQSLPFGGVK 558
+EE I LAND+ YGL +++ N RA+R A+Q+ G IN + S +YM + GG K
Sbjct: 412 EEEMISLANDTNYGLASGIWTQNIDRAMRFANQIDAGTVWINTYRSAAYMSAN---GGFK 468
Query: 559 DSGFGRFAGVEGLRACCLVKSVVED 633
+SG+GR G + +R K+VV D
Sbjct: 469 ESGYGRRGGFDVMREFSRAKNVVID 493
>sptr|Q8UKB9|Q8UKB9 Aldehyde dehydrogenase.
Length = 521
Score = 131 bits (329), Expect = 2e-29
Identities = 71/191 (37%), Positives = 107/191 (56%)
Frame = +1
Query: 25 SGQNCAGAERFYVHDDIYSAFVSQIVKTVKSISVGPPLSGRYDMGAICMIEHSEKLQNLV 204
+GQ C R VH IY ++ + ++++ VGP DMG + E+ ++ ++V
Sbjct: 316 AGQFCMTGSRLLVHSSIYELVRDRLAERLRAVKVGPASDPASDMGPLIDKENVARVDSVV 375
Query: 205 NDALDKGAEIAVRGSFGNLGEDAVDQFFPPTGLVNVDHTMKIMQEETFGPIIPIMKFSSD 384
DAL KGA + VRG G A FF P+ L D+++ I+Q+ETFGP+I I +FS +
Sbjct: 376 KDALSKGATVVVRGGPVPEGPLAKGAFFRPSMLEVFDNSLPIVQQETFGPVITIQRFSDE 435
Query: 385 EEAIKLANDSKYGLGCAVFSGNQKRAIRIASQLHCGVAAINDFASSYMCQSLPFGGVKDS 564
EA+ LAND+ YGL +V++ + R++R+A L G +ND+A Y GG K S
Sbjct: 436 REAVVLANDNDYGLSASVWTRDLDRSLRVAQALEAGSVFVNDWAKVY--DGTEEGGFKQS 493
Query: 565 GFGRFAGVEGL 597
G GR GV +
Sbjct: 494 GLGRLNGVAAI 504
>sptr|Q8GAK7|Q8GAK7 Putative succinate-semialdehyde dehydrogenase.
Length = 458
Score = 130 bits (328), Expect = 3e-29
Identities = 73/205 (35%), Positives = 109/205 (53%)
Frame = +1
Query: 10 AALQSSGQNCAGAERFYVHDDIYSAFVSQIVKTVKSISVGPPLSGRYDMGAICMIEHSEK 189
A LQ+SGQ+C A+RFYVH+D+Y F V + G PL G + +
Sbjct: 257 ARLQNSGQSCIAAKRFYVHEDVYDRFEHLFVTGMAEAVAGDPLDESTSFGPLATERGRQD 316
Query: 190 LQNLVNDALDKGAEIAVRGSFGNLGEDAVDQFFPPTGLVNVDHTMKIMQEETFGPIIPIM 369
+ LV DA +KGA + G + ++P T L V M+I +EE FGP+ +
Sbjct: 317 VHELVRDAREKGAAVQCGGEI----PEGEGWYYPATVLTGVTEDMRIYREECFGPVACLY 372
Query: 370 KFSSDEEAIKLANDSKYGLGCAVFSGNQKRAIRIASQLHCGVAAINDFASSYMCQSLPFG 549
K SS +EAI L+NDS +GL +V++ ++ A A + G IN +S+ ++PFG
Sbjct: 373 KVSSLQEAIALSNDSDFGLSSSVWTNDETEATEAARSIEAGGVFINGLTASF--PAVPFG 430
Query: 550 GVKDSGFGRFAGVEGLRACCLVKSV 624
G+KDSG+GR G+R +K+V
Sbjct: 431 GLKDSGYGRELSAYGIREFVNIKTV 455
>sptr|Q83CC3|Q83CC3 Aldehyde dehydrogenase family protein.
Length = 455
Score = 130 bits (327), Expect = 4e-29
Identities = 70/207 (33%), Positives = 118/207 (57%), Gaps = 1/207 (0%)
Frame = +1
Query: 16 LQSSGQNCAGAERFYVHDDIYSAFVSQIVKTVKSISVGPPLSGRYDMGAICMIEHSEKLQ 195
L +SGQ+C A+R + +++Y F + +++ +S +G PL ++G + + + +
Sbjct: 255 LSNSGQSCIAAKRLIIVENVYDEFKALVIEKAQSYRMGDPLKKETELGPLARSDLRDTVH 314
Query: 196 NLVNDALDKGAEIAVRGSFGNLGEDAVDQFFPPTGLVNVDHTMKIMQEETFGPIIPIMKF 375
V +++DKGA + G ++ ++PPT L V +E FGP+I +++
Sbjct: 315 GQVRESIDKGALLETGGEI----PESKGFYYPPTVLSGVRAGQPAFDDEVFGPVIALIRA 370
Query: 376 SSDEEAIKLANDSKYGLGCAVFSGNQKRAIRI-ASQLHCGVAAINDFASSYMCQSLPFGG 552
++ AI++AN+SK+GLG AVF+ + +R +I AS+L G +N F S LPFGG
Sbjct: 371 KDEKHAIEIANNSKFGLGAAVFTNDNERGEKIAASELQAGTCVVNTFVKS--DPRLPFGG 428
Query: 553 VKDSGFGRFAGVEGLRACCLVKSVVED 633
VK SG+GR EG+R+ VK+VV D
Sbjct: 429 VKHSGYGRELAAEGIRSFMNVKTVVVD 455
>sptr|Q8GFB8|Q8GFB8 Orf17.
Length = 490
Score = 130 bits (326), Expect = 5e-29
Identities = 75/203 (36%), Positives = 107/203 (52%)
Frame = +1
Query: 10 AALQSSGQNCAGAERFYVHDDIYSAFVSQIVKTVKSISVGPPLSGRYDMGAICMIEHSEK 189
A ++SGQ C A R V + IY AF ++ + V + VGP + G + +K
Sbjct: 282 AKFRNSGQTCVCANRILVQETIYDAFAERLAQAVNKLQVGPASDAKSQQGPLINQAAVDK 341
Query: 190 LQNLVNDALDKGAEIAVRGSFGNLGEDAVDQFFPPTGLVNVDHTMKIMQEETFGPIIPIM 369
++ +NDA+ +GA I G LG FF PT L +V+ +M I QEETFGP+ P+
Sbjct: 342 VEEHINDAVSRGARILAGGKPHALG----GLFFEPTVLADVNESMLISQEETFGPVAPLF 397
Query: 370 KFSSDEEAIKLANDSKYGLGCAVFSGNQKRAIRIASQLHCGVAAINDFASSYMCQSLPFG 549
KF + EAI LAND+++GL +S + R R+A L G+ IN+ S + PFG
Sbjct: 398 KFRDEAEAIHLANDTEFGLAAYFYSRDIGRIYRVAEALESGMVGINEGLISN--EVAPFG 455
Query: 550 GVKDSGFGRFAGVEGLRACCLVK 618
G+K SG GR G+ VK
Sbjct: 456 GIKQSGLGREGSRYGIEDYLEVK 478
>sptr|Q97XA5|Q97XA5 Glyceraldehyde-3-phosphate dehydrogenase, NADP
dependent (gapN-2) (EC 1.2.1.9).
Length = 470
Score = 129 bits (325), Expect = 6e-29
Identities = 73/201 (36%), Positives = 112/201 (55%)
Frame = +1
Query: 10 AALQSSGQNCAGAERFYVHDDIYSAFVSQIVKTVKSISVGPPLSGRYDMGAICMIEHSEK 189
A + +GQNC +R V ++Y FV + K++ VG PL D+G + E E
Sbjct: 267 ARFEYAGQNCNAGKRIIVRQEVYDKFVKAFNEKAKALKVGEPLDETTDVGPVINKESVEN 326
Query: 190 LQNLVNDALDKGAEIAVRGSFGNLGEDAVDQFFPPTGLVNVDHTMKIMQEETFGPIIPIM 369
L +++ DA KG + + N G ++ FFP T + N M +++ E FGPI+PI+
Sbjct: 327 LNSVLEDAKVKGGRVEIL----NRGPES-GSFFPLTMVTNPSLDMLVLKSEVFGPIVPIV 381
Query: 370 KFSSDEEAIKLANDSKYGLGCAVFSGNQKRAIRIASQLHCGVAAINDFASSYMCQSLPFG 549
SDEEAI++AN ++YGL A+F+ + RA++++ +L G IND ++ SLPFG
Sbjct: 382 SVKSDEEAIRIANSTEYGLQSAIFTNDVNRALKLSRELKFGAVIIND-STRLRWDSLPFG 440
Query: 550 GVKDSGFGRFAGVEGLRACCL 612
G K +G GR EG+R L
Sbjct: 441 GFKKTGIGR----EGVRETML 457
>sw|P71016|DHAB_BACSU Betaine aldehyde dehydrogenase (EC 1.2.1.8)
(BADH).
Length = 490
Score = 129 bits (325), Expect = 6e-29
Identities = 77/208 (37%), Positives = 113/208 (54%)
Frame = +1
Query: 1 ARGAALQSSGQNCAGAERFYVHDDIYSAFVSQIVKTVKSISVGPPLSGRYDMGAICMIEH 180
A A +GQ C+ R V D I+ F++++VK K I +G + G + EH
Sbjct: 271 ALNAVFFHAGQVCSAGSRLLVEDAIHDQFLAELVKRAKRIKLGNGFHAETESGPLISAEH 330
Query: 181 SEKLQNLVNDALDKGAEIAVRGSFGNLGEDAVDQFFPPTGLVNVDHTMKIMQEETFGPII 360
K++ V +++GA++ G E F+ PT N + M+I+QEE FGP++
Sbjct: 331 RAKVEKYVEIGIEEGAKLETGGKRPEDPELQNGFFYEPTIFSNCNSDMRIVQEEVFGPVL 390
Query: 361 PIMKFSSDEEAIKLANDSKYGLGCAVFSGNQKRAIRIASQLHCGVAAINDFASSYMCQSL 540
+ FSS+EE I+LAND+ YGL AV+S + ++ R+A++L G INDF Y Q+
Sbjct: 391 TVETFSSEEEVIELANDTIYGLAGAVWSKDIEKCERVAARLRMGTVWINDF-HPYFAQA- 448
Query: 541 PFGGVKDSGFGRFAGVEGLRACCLVKSV 624
P+GG K SGFGR G GL VK V
Sbjct: 449 PWGGYKQSGFGRELGKIGLEEYTEVKHV 476
>sptr|Q82DG6|Q82DG6 Putative aldehyde dehydrogenase.
Length = 543
Score = 129 bits (324), Expect = 8e-29
Identities = 71/213 (33%), Positives = 116/213 (54%), Gaps = 1/213 (0%)
Frame = +1
Query: 10 AALQSSGQNCAGAERFYVHDDIYSAFVSQIVKTVKSISVGPPLSGRYDMGAICMIEHSEK 189
A S+GQ C ER YVH+ + AF + +++ +G L+ DMG++ E
Sbjct: 305 ACFSSAGQLCISIERLYVHESVADAFAERFAARTRAMRLGTSLAYGADMGSLVGERQLET 364
Query: 190 LQNLVNDALDKGAEIAVRGSFGNLGEDAVDQFFPPTGLVNVDHTMKIMQEETFGPIIPIM 369
+ V++A+ KGA++ G D F+ PT L V+ M + EETFGP++ +
Sbjct: 365 VTRHVDEAVAKGAKLLAGGV---ARPDIGPYFYEPTILDGVEAPMSVCTEETFGPVVSLY 421
Query: 370 KFSSDEEAIKLANDSKYGLGCAVFSGNQKRAIRIASQLHCGVAAIND-FASSYMCQSLPF 546
+F +++EA++LAN + YGL +V++ + +R +A++L G +ND +A +Y P
Sbjct: 422 RFKTEDEAVELANSTPYGLNASVWTKDGRRGRAVAARLRTGTVNVNDGYAPAYGSVQSPM 481
Query: 547 GGVKDSGFGRFAGVEGLRACCLVKSVVEDRLWP 645
GG+KDSG GR G EG+ ++V RL P
Sbjct: 482 GGMKDSGLGRRHGSEGILKYTEAQTVAHQRLLP 514
>sptr|Q92NJ0|Q92NJ0 Putative aldehyde dehydrogenase protein (EC
1.2.1.-).
Length = 461
Score = 129 bits (324), Expect = 8e-29
Identities = 76/203 (37%), Positives = 111/203 (54%)
Frame = +1
Query: 13 ALQSSGQNCAGAERFYVHDDIYSAFVSQIVKTVKSISVGPPLSGRYDMGAICMIEHSEKL 192
A +SGQ C G ER YVH+ +Y AFV + V V + +G PL +G + + +
Sbjct: 255 ATYNSGQCCCGIERIYVHESLYDAFVEKSVAWVSNYKLGNPLDPETTLGPMANKRFAATV 314
Query: 193 QNLVNDALDKGAEIAVRGSFGNLGEDAVDQFFPPTGLVNVDHTMKIMQEETFGPIIPIMK 372
+N V DA+ KGA + +D + P LV+VDH+M+ M+EETFGP + IMK
Sbjct: 315 RNQVADAISKGARALIDPKL--FPQDDGGAYLAPQVLVDVDHSMEFMREETFGPAVGIMK 372
Query: 373 FSSDEEAIKLANDSKYGLGCAVFSGNQKRAIRIASQLHCGVAAINDFASSYMCQSLPFGG 552
SD EAI+L NDS+YGL ++++ + +RA RI L G +N + Y+ +L + G
Sbjct: 373 VKSDAEAIELMNDSRYGLTASLWTKDAERAARIGRDLETGTVFMN--RADYLDPALCWTG 430
Query: 553 VKDSGFGRFAGVEGLRACCLVKS 621
VK++G G V G KS
Sbjct: 431 VKETGRGGSLSVLGFHNLTRPKS 453
Database: /db/uniprot/tmp/swall
Posted date: Mar 5, 2004 7:45 PM
Number of letters in database: 442,889,342
Number of sequences in database: 1,395,590
Lambda K H
0.318 0.135 0.401
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 893,216,069
Number of Sequences: 1395590
Number of extensions: 18476522
Number of successful extensions: 46581
Number of sequences better than 10.0: 30
Number of HSP's better than 10.0 without gapping: 43107
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 45027
length of database: 442,889,342
effective HSP length: 125
effective length of database: 268,440,592
effective search space used: 66841707408
frameshift window, decay const: 50, 0.1
T: 12
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)

