BLASTX 2.2.6 [Apr-09-2003]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= 2643556.2.1
(1354 letters)
Database: /db/trembl-ebi/tmp/swall
1,242,019 sequences; 397,940,306 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
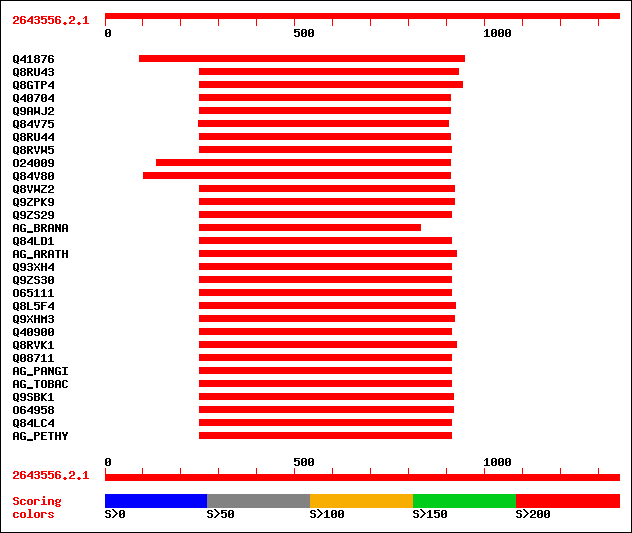
sptr|Q41876|Q41876 ZAG1 protein. 536 e-151
sptr|Q8RU43|Q8RU43 Agamous-like protein 2 HvAG2. 333 3e-90
sptr|Q8GTP4|Q8GTP4 MADS box transcription factor. 332 1e-89
sptr|Q40704|Q40704 MADS box protein. 313 3e-84
sptr|Q9AWJ2|Q9AWJ2 Putative MADS-box protein. 313 3e-84
sptr|Q84V75|Q84V75 M23 protein (Fragment). 297 3e-79
sptr|Q8RU44|Q8RU44 Agamous-like protein 1 HvAG1. 296 5e-79
sptr|Q8RVW5|Q8RVW5 MADS-box transcription factor. 286 5e-76
sptr|O24009|O24009 Agamous protein. 277 3e-73
sptr|Q84V80|Q84V80 Putative MADS-domain transcription factor (Fr... 275 1e-72
sptr|Q8VWZ2|Q8VWZ2 C-type MADS box protein. 273 3e-72
sptr|Q9ZPK9|Q9ZPK9 Agamous homolog transcription factor. 267 3e-70
sptr|Q9ZS29|Q9ZS29 MADS-box protein, GAGA2. 266 4e-70
sw|Q01540|AG_BRANA Floral homeotic protein AGAMOUS. 265 1e-69
sptr|Q84LD1|Q84LD1 MADS-box transcription factor CDM37. 265 1e-69
sw|P17839|AG_ARATH Floral homeotic protein AGAMOUS. 265 1e-69
sptr|Q93XH4|Q93XH4 MAD-box transcripion factor. 265 1e-69
sptr|Q9ZS30|Q9ZS30 MADS-box protein, GAGA1. 264 2e-69
sptr|O65111|O65111 Agamous homolog. 263 4e-69
sptr|Q8L5F4|Q8L5F4 MADS box transcription factor. 262 7e-69
sptr|Q9XHM3|Q9XHM3 Agamous homolog (Fragment). 260 3e-68
sptr|Q40900|Q40900 Agamous protein. 260 3e-68
sptr|Q8RVK1|Q8RVK1 AP3-like protein. 260 4e-68
sptr|Q08711|Q08711 Fbp6 protein. 259 8e-68
sw|Q40872|AG_PANGI Floral homeotic protein AGAMOUS (GAG2). 258 1e-67
sw|Q43585|AG_TOBAC Floral homeotic protein AGAMOUS (NAG1). 258 1e-67
sptr|Q9SBK1|Q9SBK1 Agamous-like putative transcription factor. 257 2e-67
sptr|O64958|O64958 CUM1. 257 2e-67
sptr|Q84LC4|Q84LC4 MADS-box transcriptional factor HAM45. 257 3e-67
sw|Q40885|AG_PETHY Floral homeotic protein AGAMOUS (pMADS3). 256 4e-67
>sptr|Q41876|Q41876 ZAG1 protein.
Length = 286
Score = 536 bits (1380), Expect = e-151
Identities = 272/286 (95%), Positives = 272/286 (95%)
Frame = +3
Query: 90 MHIREEEATPSTVTGIMSTLTSAGQQKLKEPIXXXXXXXXXXXXXXERNNGGRGKGKTEI 269
MHIREEEATPSTVTGIMSTLTSAGQQKLKEPI ERNNGGRGKGKTEI
Sbjct: 1 MHIREEEATPSTVTGIMSTLTSAGQQKLKEPISPGGGSASVAGSAAERNNGGRGKGKTEI 60
Query: 270 KRIENTTNRQVTFCKRRNGLLKKAYELSVLCDAEVALIVFSSRGRLYEYANNSVKGTIER 449
KRIENTTNRQVTFCKRRNGLLKKAYELSVLCDAEVALIVFSSRGRLYEYANNSVKGTIER
Sbjct: 61 KRIENTTNRQVTFCKRRNGLLKKAYELSVLCDAEVALIVFSSRGRLYEYANNSVKGTIER 120
Query: 450 YKKATSDNSSAAGTIAEVTIQHYKQESARLRQQIVNLQNSNRALIGDSITTMSHKELKHL 629
YKKATSDNSSAAGTIAEVTIQHYKQESARLRQQIVNLQNSNRALIGDSITTMSHKELKHL
Sbjct: 121 YKKATSDNSSAAGTIAEVTIQHYKQESARLRQQIVNLQNSNRALIGDSITTMSHKELKHL 180
Query: 630 ETRLDKALGKIRAKKNDVLCSEVEYMQRREMELQNDNLYLRSRVDENERAQQTANMMGAP 809
ETRLDKALGKIRAKKNDVLCSEVEYMQRREMELQNDNLYLRSRVDENERAQQTANMMGAP
Sbjct: 181 ETRLDKALGKIRAKKNDVLCSEVEYMQRREMELQNDNLYLRSRVDENERAQQTANMMGAP 240
Query: 810 STSEYQQHGFTPYDPIRSFLQFNIVQQPQFYSQQEDRKDFNDQGGR 947
STSEYQQHGFTPYDPIRSFLQFNIVQQPQFYSQQEDRKDFNDQGGR
Sbjct: 241 STSEYQQHGFTPYDPIRSFLQFNIVQQPQFYSQQEDRKDFNDQGGR 286
>sptr|Q8RU43|Q8RU43 Agamous-like protein 2 HvAG2.
Length = 232
Score = 333 bits (854), Expect = 3e-90
Identities = 170/229 (74%), Positives = 194/229 (84%), Gaps = 1/229 (0%)
Frame = +3
Query: 249 GKGKTEIKRIENTTNRQVTFCKRRNGLLKKAYELSVLCDAEVALIVFSSRGRLYEYANNS 428
G+G+ EIKRIENTTNRQ+TFCKRRNGLLKKAYELSVLCDAEVAL+VFSSRGRLYEY+NNS
Sbjct: 2 GRGRIEIKRIENTTNRQLTFCKRRNGLLKKAYELSVLCDAEVALVVFSSRGRLYEYSNNS 61
Query: 429 VKGTIERYKKATSDNSSAAGTIAEVTIQHYKQESARLRQQIVNLQNSNRALIGDSITTMS 608
VK TIERYKKATSD SSA GT+AE+ QHY+QESA+LRQQI LQNSNR LIGD++ TMS
Sbjct: 62 VKATIERYKKATSDTSSA-GTVAEINAQHYQQESAKLRQQITTLQNSNRTLIGDTMATMS 120
Query: 609 HKELKHLETRLDKALGKIRAKKNDVLCSEVEYMQRREMELQNDNLYLRSRVDENERA-QQ 785
H++LK LE RLDK LGKIRA+KN++L +E+EYMQRREMELQN+N YLR +V E ER QQ
Sbjct: 121 HRDLKQLEGRLDKGLGKIRARKNELLSAEIEYMQRREMELQNNNFYLREKVAETERGQQQ 180
Query: 786 TANMMGAPSTSEYQQHGFTPYDPIRSFLQFNIVQQPQFYSQQEDRKDFN 932
T NMMGA STS DP R+FLQFNI+QQPQ+Y+QQEDRK N
Sbjct: 181 TLNMMGAASTSNEYDQNMIQCDP-RTFLQFNIMQQPQYYTQQEDRKTLN 228
>sptr|Q8GTP4|Q8GTP4 MADS box transcription factor.
Length = 254
Score = 332 bits (850), Expect = 1e-89
Identities = 171/233 (73%), Positives = 195/233 (83%), Gaps = 2/233 (0%)
Frame = +3
Query: 249 GKGKTEIKRIENTTNRQVTFCKRRNGLLKKAYELSVLCDAEVALIVFSSRGRLYEYANNS 428
G+G+ EIKRIENTTNRQVTFCKRRNGLLKKAYELSVLCDAEVALIVFS RGRLYEY+NNS
Sbjct: 23 GRGRIEIKRIENTTNRQVTFCKRRNGLLKKAYELSVLCDAEVALIVFSGRGRLYEYSNNS 82
Query: 429 VKGTIERYKKATSDNSSAAGTIAEVTIQHYKQESARLRQQIVNLQNSNRALIGDSITTMS 608
VK TIERYKKATSD SSA GT+AE+ QHY+QESA+L+QQI LQNSNR LIGD++ TMS
Sbjct: 83 VKATIERYKKATSDTSSA-GTVAEINAQHYQQESAKLKQQITTLQNSNRTLIGDTMATMS 141
Query: 609 HKELKHLETRLDKALGKIRAKKNDVLCSEVEYMQRREMELQNDNLYLRSRVDENERA-QQ 785
H++LK LE RLDK LGKIRA+KN++LC+E+EYMQRREMELQN+N +LR +V E ER QQ
Sbjct: 142 HRDLKQLEGRLDKGLGKIRARKNELLCAEIEYMQRREMELQNNNFFLREKVAETERGQQQ 201
Query: 786 TANMMGAPSTSEYQQHGFTPYDPIRSFLQFNIV-QQPQFYSQQEDRKDFNDQG 941
T NMMGA STS + DP R+FLQFN + QQPQ+YSQQEDRK N G
Sbjct: 202 TLNMMGAASTSNEYEQNMIHCDP-RTFLQFNFMQQQPQYYSQQEDRKSLNSVG 253
>sptr|Q40704|Q40704 MADS box protein.
Length = 236
Score = 313 bits (803), Expect = 3e-84
Identities = 161/222 (72%), Positives = 189/222 (85%), Gaps = 1/222 (0%)
Frame = +3
Query: 249 GKGKTEIKRIENTTNRQVTFCKRRNGLLKKAYELSVLCDAEVALIVFSSRGRLYEYANNS 428
G+GK EIKRIENTTNRQVTFCKRRNGLLKKAYELSVLCDAEVALIVFSSRGRLYEYANNS
Sbjct: 2 GRGKIEIKRIENTTNRQVTFCKRRNGLLKKAYELSVLCDAEVALIVFSSRGRLYEYANNS 61
Query: 429 VKGTIERYKKATSDNSSAAGTIAEVTIQHYKQESARLRQQIVNLQNSN-RALIGDSITTM 605
VK T+ERYKKA SD S+ +GT+AEV QHY+QES++LRQQI +LQN+N R ++GDSI TM
Sbjct: 62 VKSTVERYKKANSDTSN-SGTVAEVNAQHYQQESSKLRQQISSLQNANSRTIVGDSINTM 120
Query: 606 SHKELKHLETRLDKALGKIRAKKNDVLCSEVEYMQRREMELQNDNLYLRSRVDENERAQQ 785
S ++LK +E RL+K + KIRA+KN++L +EVEYMQ+RE+ELQNDN+YLRS+V ENER QQ
Sbjct: 121 SLRDLKQVENRLEKGIAKIRARKNELLYAEVEYMQKREVELQNDNMYLRSKVVENERGQQ 180
Query: 786 TANMMGAPSTSEYQQHGFTPYDPIRSFLQFNIVQQPQFYSQQ 911
NMMGA STSEY PYD R+FLQ NI+QQPQ Y+ Q
Sbjct: 181 PLNMMGAASTSEYDHMVNNPYDS-RNFLQVNIMQQPQHYAHQ 221
>sptr|Q9AWJ2|Q9AWJ2 Putative MADS-box protein.
Length = 247
Score = 313 bits (803), Expect = 3e-84
Identities = 161/222 (72%), Positives = 189/222 (85%), Gaps = 1/222 (0%)
Frame = +3
Query: 249 GKGKTEIKRIENTTNRQVTFCKRRNGLLKKAYELSVLCDAEVALIVFSSRGRLYEYANNS 428
G+GK EIKRIENTTNRQVTFCKRRNGLLKKAYELSVLCDAEVALIVFSSRGRLYEYANNS
Sbjct: 2 GRGKIEIKRIENTTNRQVTFCKRRNGLLKKAYELSVLCDAEVALIVFSSRGRLYEYANNS 61
Query: 429 VKGTIERYKKATSDNSSAAGTIAEVTIQHYKQESARLRQQIVNLQNSN-RALIGDSITTM 605
VK T+ERYKKA SD S+ +GT+AEV QHY+QES++LRQQI +LQN+N R ++GDSI TM
Sbjct: 62 VKSTVERYKKANSDTSN-SGTVAEVNAQHYQQESSKLRQQISSLQNANSRTIVGDSINTM 120
Query: 606 SHKELKHLETRLDKALGKIRAKKNDVLCSEVEYMQRREMELQNDNLYLRSRVDENERAQQ 785
S ++LK +E RL+K + KIRA+KN++L +EVEYMQ+RE+ELQNDN+YLRS+V ENER QQ
Sbjct: 121 SLRDLKQVENRLEKGIAKIRARKNELLYAEVEYMQKREVELQNDNMYLRSKVVENERGQQ 180
Query: 786 TANMMGAPSTSEYQQHGFTPYDPIRSFLQFNIVQQPQFYSQQ 911
NMMGA STSEY PYD R+FLQ NI+QQPQ Y+ Q
Sbjct: 181 PLNMMGAASTSEYDHMVNNPYDS-RNFLQVNIMQQPQHYAHQ 221
>sptr|Q84V75|Q84V75 M23 protein (Fragment).
Length = 304
Score = 297 bits (760), Expect = 3e-79
Identities = 152/221 (68%), Positives = 183/221 (82%), Gaps = 1/221 (0%)
Frame = +3
Query: 246 RGKGKTEIKRIENTTNRQVTFCKRRNGLLKKAYELSVLCDAEVALIVFSSRGRLYEYANN 425
+G+GK EIKRIENTTNRQVTFCKRRNGLLKKAYELSVLCDAEVAL+VFSSRGRLYEYANN
Sbjct: 58 QGRGKIEIKRIENTTNRQVTFCKRRNGLLKKAYELSVLCDAEVALVVFSSRGRLYEYANN 117
Query: 426 SVKGTIERYKKATSDNSSAAGTIAEVTIQHYKQESARLRQQIVNLQNSNRALIGDSITTM 605
SVK TIERYKKA SD S+ +GT+AEV QHY+QES++LRQ I +LQN+NR ++GDSI TM
Sbjct: 118 SVKSTIERYKKANSDTSN-SGTVAEVNAQHYQQESSKLRQAIDSLQNANRTIVGDSIHTM 176
Query: 606 SHKELKHLETRLDKALGKIRAKKNDVLCSEVEYMQRREMELQNDNLYLRSRVDE-NERAQ 782
+ELK +E +L+KA+ KIRA+KN++L +EVEYMQ+REM+LQ DN+YLRS++ E NE Q
Sbjct: 177 GLRELKQMEGKLEKAINKIRARKNELLYAEVEYMQKREMDLQTDNMYLRSKIAENNETGQ 236
Query: 783 QTANMMGAPSTSEYQQHGFTPYDPIRSFLQFNIVQQPQFYS 905
NM+G PSTSEY P+ R+FLQ N+ QQPQ YS
Sbjct: 237 PPMNMIGLPSTSEYDH--MAPFVDSRNFLQVNMQQQPQHYS 275
>sptr|Q8RU44|Q8RU44 Agamous-like protein 1 HvAG1.
Length = 234
Score = 296 bits (758), Expect = 5e-79
Identities = 154/222 (69%), Positives = 186/222 (83%), Gaps = 1/222 (0%)
Frame = +3
Query: 249 GKGKTEIKRIENTTNRQVTFCKRRNGLLKKAYELSVLCDAEVALIVFSSRGRLYEYANNS 428
G+G+ EIKRIENTTNRQVTFCKRRNGLLKKAYELSVLCDAEVAL+VFSSRGRLYEY+NNS
Sbjct: 2 GRGRIEIKRIENTTNRQVTFCKRRNGLLKKAYELSVLCDAEVALVVFSSRGRLYEYSNNS 61
Query: 429 VKGTIERYKKATSDNSSAAGTIAEVTIQHYKQESARLRQQIVNLQNSN-RALIGDSITTM 605
VK TIERYKKA SD S+ +GT+AEV Q+Y+QES++LRQQI +LQNSN R+L+ DS++TM
Sbjct: 62 VKATIERYKKANSDTSN-SGTVAEVNAQYYQQESSKLRQQISSLQNSNSRSLVRDSVSTM 120
Query: 606 SHKELKHLETRLDKALGKIRAKKNDVLCSEVEYMQRREMELQNDNLYLRSRVDENERAQQ 785
+ ++LK LE RL+K + KIRA+KN+++ +EVEYMQ+REMEL NDN+YLRS+V ENER QQ
Sbjct: 121 TLRDLKQLEGRLEKGIAKIRARKNELMYAEVEYMQKREMELHNDNIYLRSKVSENERGQQ 180
Query: 786 TANMMGAPSTSEYQQHGFTPYDPIRSFLQFNIVQQPQFYSQQ 911
NMM + STS H PYD R+FLQ N +QQ Q YSQQ
Sbjct: 181 PMNMMASGSTSSEYDHMVAPYDS-RNFLQVN-MQQQQHYSQQ 220
>sptr|Q8RVW5|Q8RVW5 MADS-box transcription factor.
Length = 239
Score = 286 bits (732), Expect = 5e-76
Identities = 149/224 (66%), Positives = 185/224 (82%), Gaps = 2/224 (0%)
Frame = +3
Query: 249 GKGKTEIKRIENTTNRQVTFCKRRNGLLKKAYELSVLCDAEVALIVFSSRGRLYEYANNS 428
G+GK EIKRIENTTNRQVTFCKRRNGLLKKAYELSVLCDAEVALI+FS+RGRLYEYANNS
Sbjct: 13 GRGKIEIKRIENTTNRQVTFCKRRNGLLKKAYELSVLCDAEVALIIFSTRGRLYEYANNS 72
Query: 429 VKGTIERYKKATSDNSSAAGTIAEVTIQHYKQESARLRQQIVNLQNSNRALIGDSITTMS 608
VKGTIERYKKA++DNS+ G+I+E Q+Y+QE+ +LRQQI NLQNSNR L+GD++TTMS
Sbjct: 73 VKGTIERYKKASTDNSN-TGSISEANSQYYQQEATKLRQQITNLQNSNRNLLGDALTTMS 131
Query: 609 HKELKHLETRLDKALGKIRAKKNDVLCSEVEYMQRREMELQNDNLYLRSRVDENERAQQT 788
++LK LETRL+K + KIRAKKN++L +E++YMQ+REMELQ DN++LR+++ +NERAQQ
Sbjct: 132 LRDLKQLETRLEKGINKIRAKKNELLHAEIDYMQKREMELQTDNMFLRNKISDNERAQQQ 191
Query: 789 ANMMG-APSTS-EYQQHGFTPYDPIRSFLQFNIVQQPQFYSQQE 914
M PSTS EY+ P+D RSFL N++ YS Q+
Sbjct: 192 HQHMSILPSTSTEYEV--MPPFDS-RSFLHVNLMDPNDRYSHQQ 232
>sptr|O24009|O24009 Agamous protein.
Length = 259
Score = 277 bits (708), Expect = 3e-73
Identities = 156/262 (59%), Positives = 194/262 (74%), Gaps = 3/262 (1%)
Frame = +3
Query: 135 IMSTLTSAGQQKLKEPIXXXXXXXXXXXXXXERNNGGRGKGKTEIKRIENTTNRQVTFCK 314
+M+ L+ K+KE + + G+G+GK EIKRIENTTNRQVTFCK
Sbjct: 4 MMTDLSCGPSSKVKEQVAAAPTGSG--------DRQGQGRGKIEIKRIENTTNRQVTFCK 55
Query: 315 RRNGLLKKAYELSVLCDAEVALIVFSSRGRLYEYANNSVKGTIERYKKATSDNSSAAGTI 494
RRNGLLKKAYELSVLCDAEVAL+VFSSRGRLYEYANNSVK TIERYKKA SD SS +GT+
Sbjct: 56 RRNGLLKKAYELSVLCDAEVALVVFSSRGRLYEYANNSVKSTIERYKKANSD-SSNSGTV 114
Query: 495 AEVTIQHYKQESARLRQQIVNLQNSN-RALIGDSITTMSHKELKHLETRLDKALGKIRAK 671
AEV Q+Y+QES++LRQ I +LQN+N R ++GDSI TM ++LK +E +L+KA+ KIRA+
Sbjct: 115 AEVNAQYYQQESSKLRQMIHSLQNANTRNIVGDSIHTMGLRDLKQMEGKLEKAIIKIRAR 174
Query: 672 KNDVLCSEVEYMQRREMELQNDNLYLRSRVDE-NERAQQTANM-MGAPSTSEYQQHGFTP 845
KN++L +EV+YMQ+REM+LQ DN+YLRS++ E NE Q +M MGAP TSEY P
Sbjct: 175 KNELLYAEVDYMQKREMDLQTDNMYLRSKIAESNETGQPAMHMTMGAPPTSEYDH--MAP 232
Query: 846 YDPIRSFLQFNIVQQPQFYSQQ 911
+D R+FLQ V PQ YS Q
Sbjct: 233 FDS-RNFLQ---VSMPQHYSHQ 250
>sptr|Q84V80|Q84V80 Putative MADS-domain transcription factor
(Fragment).
Length = 273
Score = 275 bits (703), Expect = 1e-72
Identities = 157/276 (56%), Positives = 197/276 (71%), Gaps = 6/276 (2%)
Frame = +3
Query: 102 EEEATPS---TVTGIMSTLTSAGQQKLKEPIXXXXXXXXXXXXXXERNNGGRGKGKTEIK 272
+ EA P + +M+ L+ K+KE + + G+G+GK EIK
Sbjct: 4 DREALPHHHHPMLNMMTDLSCGPSSKVKEQVAAAPTGSG--------DRQGQGRGKIEIK 55
Query: 273 RIENTTNRQVTFCKRRNGLLKKAYELSVLCDAEVALIVFSSRGRLYEYANNSVKGTIERY 452
RIENTTNRQVTFCKRRNGLLKKAYELSVLCDAEVAL+VFSSRGRLYEYANNSVK TIERY
Sbjct: 56 RIENTTNRQVTFCKRRNGLLKKAYELSVLCDAEVALVVFSSRGRLYEYANNSVKSTIERY 115
Query: 453 KKATSDNSSAAGTIAEVTIQHYKQESARLRQQIVNLQNSN-RALIGDSITTMSHKELKHL 629
KKA SD SS +GT+AEV Q+Y+QES++LRQ I +LQN+N R ++GDSI TM ++LK +
Sbjct: 116 KKANSD-SSNSGTVAEVNAQYYQQESSKLRQMIHSLQNANTRNIVGDSIHTMGLRDLKQM 174
Query: 630 ETRLDKALGKIRAKKNDVLCSEVEYMQRREMELQNDNLYLRSRVDE-NERAQQTANMMGA 806
E +L+KA+ KIRA+KN++L +EV+YMQ+REM+LQ DN+YL S++ E NE Q +MMG
Sbjct: 175 EGKLEKAISKIRARKNELLYAEVDYMQKREMDLQTDNMYLTSKIAESNETGQPAMHMMGV 234
Query: 807 -PSTSEYQQHGFTPYDPIRSFLQFNIVQQPQFYSQQ 911
P TSEY P+D R+FLQ V PQ YS Q
Sbjct: 235 PPPTSEYDH--MAPFDS-RNFLQ---VSMPQHYSHQ 264
>sptr|Q8VWZ2|Q8VWZ2 C-type MADS box protein.
Length = 245
Score = 273 bits (699), Expect = 3e-72
Identities = 137/224 (61%), Positives = 180/224 (80%)
Frame = +3
Query: 249 GKGKTEIKRIENTTNRQVTFCKRRNGLLKKAYELSVLCDAEVALIVFSSRGRLYEYANNS 428
G+GK EIKRIENTTNRQVTFCKRRNGLLKKAYELSVLCDAEVALIVFS+RGRLYEYANNS
Sbjct: 18 GRGKIEIKRIENTTNRQVTFCKRRNGLLKKAYELSVLCDAEVALIVFSNRGRLYEYANNS 77
Query: 429 VKGTIERYKKATSDNSSAAGTIAEVTIQHYKQESARLRQQIVNLQNSNRALIGDSITTMS 608
VKGTIERYKKA++D SS G+++E + Q+Y+QE+A+LR +IV LQN NR ++GD++ +MS
Sbjct: 78 VKGTIERYKKASAD-SSNTGSVSEASTQYYQQEAAKLRARIVKLQNDNRNMMGDALNSMS 136
Query: 609 HKELKHLETRLDKALGKIRAKKNDVLCSEVEYMQRREMELQNDNLYLRSRVDENERAQQT 788
K+LK LE +L+KA+ +IR+KKN++L +E+EYMQ+RE++L N+N LR+++ ENERA +T
Sbjct: 137 VKDLKSLENKLEKAISRIRSKKNELLFAEIEYMQKRELDLHNNNQLLRAKIAENERASRT 196
Query: 789 ANMMGAPSTSEYQQHGFTPYDPIRSFLQFNIVQQPQFYSQQEDR 920
N+M TS Y PYD R++ Q N +Q Y+ + D+
Sbjct: 197 LNVMAGGGTSSYDILQSQPYDS-RNYFQVNALQPNHQYNPRHDQ 239
>sptr|Q9ZPK9|Q9ZPK9 Agamous homolog transcription factor.
Length = 228
Score = 267 bits (682), Expect = 3e-70
Identities = 141/227 (62%), Positives = 185/227 (81%), Gaps = 3/227 (1%)
Frame = +3
Query: 249 GKGKTEIKRIENTTNRQVTFCKRRNGLLKKAYELSVLCDAEVALIVFSSRGRLYEYAN-N 425
G+GK EIKRIENTT+RQVTFCKRRNGLLKKAYELSVLCDAEVALIVFS+RGRLYEY+N N
Sbjct: 2 GRGKIEIKRIENTTSRQVTFCKRRNGLLKKAYELSVLCDAEVALIVFSTRGRLYEYSNSN 61
Query: 426 SVKGTIERYKKATSDNSSAAGTIAEVTIQHYKQESARLRQQIVNLQNSNRALIGDSITTM 605
SVK TIERYKKA +D ++ GT++E Q+Y+QE+ +LRQQI NLQN+NR L+G+S++TM
Sbjct: 62 SVKTTIERYKKACTDTTN-TGTVSEANSQYYQQEATKLRQQITNLQNTNRTLMGESLSTM 120
Query: 606 SHKELKHLETRLDKALGKIRAKKNDVLCSEVEYMQRREMELQNDNLYLRSRVDENERAQQ 785
S +ELK LE RL++ + KIR KKN++L +E+EYMQ+RE E+ NDN+YLR+++ ENERAQQ
Sbjct: 121 SLRELKQLEGRLERGINKIRTKKNELLSAEIEYMQKREAEMHNDNMYLRNKIAENERAQQ 180
Query: 786 TANMMGAPST-SEYQQHGFTPYDPIRSFLQFNIVQ-QPQFYSQQEDR 920
NM+ PST +EY+ G +D R+FLQ ++++ YS+Q+ +
Sbjct: 181 QMNML--PSTATEYE--GIPQFDS-RNFLQVSLMEPNNHHYSRQQQQ 222
>sptr|Q9ZS29|Q9ZS29 MADS-box protein, GAGA2.
Length = 246
Score = 266 bits (681), Expect = 4e-70
Identities = 142/228 (62%), Positives = 179/228 (78%), Gaps = 6/228 (2%)
Frame = +3
Query: 249 GKGKTEIKRIENTTNRQVTFCKRRNGLLKKAYELSVLCDAEVALIVFSSRGRLYEYANNS 428
G+GK EIKRIENTTNRQVTFCKRRNGLLKKAYELSVLCDAEVALIVFSSRGRLYEYANNS
Sbjct: 17 GRGKIEIKRIENTTNRQVTFCKRRNGLLKKAYELSVLCDAEVALIVFSSRGRLYEYANNS 76
Query: 429 VKGTIERYKKATSDNSSAAGTIAEVTIQHYKQESARLRQQIVNLQNSNRA----LIGDSI 596
VKGTI+RYKKA D S +G++AE Q Y+QE+A+LRQQI NLQN NR ++G+S+
Sbjct: 77 VKGTIDRYKKACLDPPS-SGSVAEANAQFYQQEAAKLRQQIANLQNQNRQFYRNIMGESL 135
Query: 597 TTMSHKELKHLETRLDKALGKIRAKKNDVLCSEVEYMQRREMELQNDNLYLRSRVDENER 776
M K+LK+LE++L+K +GKIR+KKN++L +E+EYMQ+RE EL N N +LRS++ ENER
Sbjct: 136 GNMPAKDLKNLESKLEKGIGKIRSKKNEILFAEIEYMQKRENELHNSNQFLRSKIAENER 195
Query: 777 AQQTANMMGAPSTSEYQQHGFTPYDPI--RSFLQFNIVQQPQFYSQQE 914
AQQ ++M P +S+Y+ P+ P R++LQ N +Q YS Q+
Sbjct: 196 AQQHMSLM--PGSSDYEL--VAPHQPFDGRNYLQVNDLQPNNNYSCQD 239
>sw|Q01540|AG_BRANA Floral homeotic protein AGAMOUS.
Length = 252
Score = 265 bits (677), Expect = 1e-69
Identities = 131/194 (67%), Positives = 169/194 (87%)
Frame = +3
Query: 249 GKGKTEIKRIENTTNRQVTFCKRRNGLLKKAYELSVLCDAEVALIVFSSRGRLYEYANNS 428
G+GK EIKRIENTTNRQVTFCKRRNGLLKKAYELSVLCDAEVALIVFSSRGRLYEY+NNS
Sbjct: 18 GRGKIEIKRIENTTNRQVTFCKRRNGLLKKAYELSVLCDAEVALIVFSSRGRLYEYSNNS 77
Query: 429 VKGTIERYKKATSDNSSAAGTIAEVTIQHYKQESARLRQQIVNLQNSNRALIGDSITTMS 608
VKGTIERYKKA SDNS+ G++AE+ Q+Y+QESA+LRQQI+++QNSNR L+G++I +MS
Sbjct: 78 VKGTIERYKKAISDNSN-TGSVAEINAQYYQQESAKLRQQIISIQNSNRQLMGETIGSMS 136
Query: 609 HKELKHLETRLDKALGKIRAKKNDVLCSEVEYMQRREMELQNDNLYLRSRVDENERAQQT 788
KEL++LE RLD+++ +IR+KKN++L +E++YMQ+RE++L NDN LR+++ ENER +
Sbjct: 137 PKELRNLEGRLDRSVNRIRSKKNELLFAEIDYMQKREVDLHNDNQLLRAKIAENERNNPS 196
Query: 789 ANMMGAPSTSEYQQ 830
++M P S Y+Q
Sbjct: 197 MSLM--PGGSNYEQ 208
>sptr|Q84LD1|Q84LD1 MADS-box transcription factor CDM37.
Length = 265
Score = 265 bits (677), Expect = 1e-69
Identities = 138/228 (60%), Positives = 180/228 (78%), Gaps = 6/228 (2%)
Frame = +3
Query: 249 GKGKTEIKRIENTTNRQVTFCKRRNGLLKKAYELSVLCDAEVALIVFSSRGRLYEYANNS 428
GKGK EIKRIENTTNRQVTFCKRRNGLLKKAYELSVLCDAEVALIVFSSRGRLYEYANNS
Sbjct: 34 GKGKIEIKRIENTTNRQVTFCKRRNGLLKKAYELSVLCDAEVALIVFSSRGRLYEYANNS 93
Query: 429 VKGTIERYKKATSDNSSAAGTIAEVTIQHYKQESARLRQQIVNLQNSNRA----LIGDSI 596
V+GTI+RYKKA D S +G+++E Q+Y++ES +LR QI NLQN NR ++G+S+
Sbjct: 94 VRGTIDRYKKACLDPPS-SGSVSEANAQYYQEESGKLRSQIANLQNQNRQFYRNIMGESL 152
Query: 597 TTMSHKELKHLETRLDKALGKIRAKKNDVLCSEVEYMQRREMELQNDNLYLRSRVDENER 776
T M K+LK+LET+L+KA+ +IR+KKN++L +E+EYMQ+RE+EL N+N +LR+++ ENER
Sbjct: 153 TDMPMKDLKNLETKLEKAITRIRSKKNELLFAEIEYMQKRELELHNNNQFLRAKIAENER 212
Query: 777 AQQTANMMGAPSTSEYQQHGFTPYDPI--RSFLQFNIVQQPQFYSQQE 914
+ Q +M P +S+Y+ TP+ P R++LQ N +Q YS Q+
Sbjct: 213 SAQQQHMSLMPGSSDYEL--VTPHQPFDGRNYLQSNEMQPSNDYSCQD 258
>sw|P17839|AG_ARATH Floral homeotic protein AGAMOUS.
Length = 252
Score = 265 bits (676), Expect = 1e-69
Identities = 138/232 (59%), Positives = 182/232 (78%), Gaps = 6/232 (2%)
Frame = +3
Query: 249 GKGKTEIKRIENTTNRQVTFCKRRNGLLKKAYELSVLCDAEVALIVFSSRGRLYEYANNS 428
G+GK EIKRIENTTNRQVTFCKRRNGLLKKAYELSVLCDAEVALIVFSSRGRLYEY+NNS
Sbjct: 18 GRGKIEIKRIENTTNRQVTFCKRRNGLLKKAYELSVLCDAEVALIVFSSRGRLYEYSNNS 77
Query: 429 VKGTIERYKKATSDNSSAAGTIAEVTIQHYKQESARLRQQIVNLQNSNRALIGDSITTMS 608
VKGTIERYKKA SDNS+ G++AE+ Q+Y+QESA+LRQQI+++QNSNR L+G++I +MS
Sbjct: 78 VKGTIERYKKAISDNSN-TGSVAEINAQYYQQESAKLRQQIISIQNSNRQLMGETIGSMS 136
Query: 609 HKELKHLETRLDKALGKIRAKKNDVLCSEVEYMQRREMELQNDNLYLRSRVDENERAQQT 788
KEL++LE RL++++ +IR+KKN++L SE++YMQ+RE++L NDN LR+++ ENER +
Sbjct: 137 PKELRNLEGRLERSITRIRSKKNELLFSEIDYMQKREVDLHNDNQILRAKIAENERNNPS 196
Query: 789 ANMMGAPSTSEYQ------QHGFTPYDPIRSFLQFNIVQQPQFYSQQEDRKD 926
++M P S Y+ Q P+D R++ Q +Q + R+D
Sbjct: 197 ISLM--PGGSNYEQLMPPPQTQSQPFDS-RNYFQVAALQPNNHHYSSAGRQD 245
>sptr|Q93XH4|Q93XH4 MAD-box transcripion factor.
Length = 225
Score = 265 bits (676), Expect = 1e-69
Identities = 134/222 (60%), Positives = 179/222 (80%)
Frame = +3
Query: 249 GKGKTEIKRIENTTNRQVTFCKRRNGLLKKAYELSVLCDAEVALIVFSSRGRLYEYANNS 428
G+GK EIKRIENTTNRQVTFCKRRNGLLKKAYELSVLCDAEVALIVFSSRGRLYEYANNS
Sbjct: 2 GRGKIEIKRIENTTNRQVTFCKRRNGLLKKAYELSVLCDAEVALIVFSSRGRLYEYANNS 61
Query: 429 VKGTIERYKKATSDNSSAAGTIAEVTIQHYKQESARLRQQIVNLQNSNRALIGDSITTMS 608
V+ TIERYKK SD SS G+++E Q Y+QE+++LR+QI ++QN NR ++G+++++++
Sbjct: 62 VRTTIERYKKVCSD-SSNTGSVSEANAQFYQQEASKLRRQIRDIQNLNRHILGEALSSLN 120
Query: 609 HKELKHLETRLDKALGKIRAKKNDVLCSEVEYMQRREMELQNDNLYLRSRVDENERAQQT 788
KELK+LETRL+K + +IR+KKN++L +E+EYMQ+RE+ELQN NL+LR+++ ENERAQQ
Sbjct: 121 FKELKNLETRLEKGISRIRSKKNELLFAEIEYMQKREIELQNSNLFLRAQIAENERAQQQ 180
Query: 789 ANMMGAPSTSEYQQHGFTPYDPIRSFLQFNIVQQPQFYSQQE 914
N+M S+Y+ PYD ++ L N++ YS+ +
Sbjct: 181 MNLMPG---SQYESVPQQPYDS-QNLLPVNLLDPNHHYSRHD 218
>sptr|Q9ZS30|Q9ZS30 MADS-box protein, GAGA1.
Length = 264
Score = 264 bits (674), Expect = 2e-69
Identities = 142/228 (62%), Positives = 179/228 (78%), Gaps = 6/228 (2%)
Frame = +3
Query: 249 GKGKTEIKRIENTTNRQVTFCKRRNGLLKKAYELSVLCDAEVALIVFSSRGRLYEYANNS 428
GKGK EIKRIENTTNRQVTFCKRRNGLLKKAYELSVLCDAEVALIVFSSRGRLYEYANNS
Sbjct: 34 GKGKIEIKRIENTTNRQVTFCKRRNGLLKKAYELSVLCDAEVALIVFSSRGRLYEYANNS 93
Query: 429 VKGTIERYKKATSDNSSAAGTIAEVTIQHYKQESARLRQQIVNLQNSNRA----LIGDSI 596
VKGTI++YKKA D +GT+AE Q+Y+QE+A+LRQQI NLQN NR ++G+S+
Sbjct: 94 VKGTIDKYKKACLD-PPTSGTVAEANTQYYQQEAAKLRQQIANLQNQNRQFYRNIMGESL 152
Query: 597 TTMSHKELKHLETRLDKALGKIRAKKNDVLCSEVEYMQRREMELQNDNLYLRSRVDENER 776
M K+LK+LE +L+KA+ +IRAKKN++L +E+EYMQ+RE+EL N N +LR+++ ENER
Sbjct: 153 GDMPVKDLKNLEGKLEKAISRIRAKKNELLFAEIEYMQKRELELHNSNQFLRAKIVENER 212
Query: 777 AQQTANMMGAPSTSEYQQHGFTPYDPI--RSFLQFNIVQQPQFYSQQE 914
AQQ +M P +S+Y+ TP+ P R++LQ N +Q YS Q+
Sbjct: 213 AQQ-HHMSLMPGSSDYEL--VTPHQPFDGRNYLQTNDLQPNNDYSCQD 257
>sptr|O65111|O65111 Agamous homolog.
Length = 241
Score = 263 bits (672), Expect = 4e-69
Identities = 133/222 (59%), Positives = 180/222 (81%)
Frame = +3
Query: 249 GKGKTEIKRIENTTNRQVTFCKRRNGLLKKAYELSVLCDAEVALIVFSSRGRLYEYANNS 428
G+GK EIKRIENTTNRQVTFCKRR+GLLKKAYELSVLCDAEVALIVFSSRGRLYEY+N+S
Sbjct: 17 GRGKVEIKRIENTTNRQVTFCKRRSGLLKKAYELSVLCDAEVALIVFSSRGRLYEYSNDS 76
Query: 429 VKGTIERYKKATSDNSSAAGTIAEVTIQHYKQESARLRQQIVNLQNSNRALIGDSITTMS 608
VK TIERYKKA++D SS G+++E Q+Y+QE+A+LR QI NLQNSNR ++G++++++S
Sbjct: 77 VKSTIERYKKASAD-SSNTGSVSEANAQYYQQEAAKLRSQIGNLQNSNRHMLGEALSSLS 135
Query: 609 HKELKHLETRLDKALGKIRAKKNDVLCSEVEYMQRREMELQNDNLYLRSRVDENERAQQT 788
KELK LE RL+K + +IR+KKN++L +E+EYMQ+RE++L N+N LR+++ ENER +Q+
Sbjct: 136 VKELKSLEIRLEKGISRIRSKKNELLFAEIEYMQKREVDLHNNNQLLRAKISENERKRQS 195
Query: 789 ANMMGAPSTSEYQQHGFTPYDPIRSFLQFNIVQQPQFYSQQE 914
N+M P ++++ PYD R++ Q N +Q YS Q+
Sbjct: 196 MNLM--PGGADFEIVQSQPYDS-RNYSQVNGLQPASHYSHQD 234
>sptr|Q8L5F4|Q8L5F4 MADS box transcription factor.
Length = 255
Score = 262 bits (670), Expect = 7e-69
Identities = 137/229 (59%), Positives = 180/229 (78%), Gaps = 4/229 (1%)
Frame = +3
Query: 249 GKGKTEIKRIENTTNRQVTFCKRRNGLLKKAYELSVLCDAEVALIVFSSRGRLYEYANNS 428
G+GK EIKRIENTTNRQVTFCKRRNGLLKKAYELSVLCDAEVALIVFS+RGRLYEYANNS
Sbjct: 19 GRGKIEIKRIENTTNRQVTFCKRRNGLLKKAYELSVLCDAEVALIVFSNRGRLYEYANNS 78
Query: 429 VKGTIERYKKATSDNSSAAGTIAEVTIQHYKQESARLRQQIVNLQNSNRALIGDSITTMS 608
V+GTIERYKKA SD + A T++E Q+Y++E+ARLRQQI NLQNSNR L+G+++ +
Sbjct: 79 VRGTIERYKKANSDTPNTA-TVSEANTQYYQKEAARLRQQISNLQNSNRHLMGEALGAVP 137
Query: 609 HKELKHLETRLDKALGKIRAKKNDVLCSEVEYMQRREMELQNDNLYLRSRVDENERAQQT 788
KELK LET+L L ++R+KKN++L +E+E+M++RE++L N+N YLR+++ ENERAQQ
Sbjct: 138 AKELKGLETKLQNGLSRVRSKKNELLFAEIEFMRKREIDLHNNNQYLRAKISENERAQQQ 197
Query: 789 ANMM-GAPSTSEYQQHGFTPYDPI--RSFLQFNIVQQPQF-YSQQEDRK 923
++M GA +SE + P++ R++LQ N +Q YS +D +
Sbjct: 198 MSLMPGASGSSEQYRDVGQPHESFDARNYLQVNGLQPNNANYSSHQDHQ 246
>sptr|Q9XHM3|Q9XHM3 Agamous homolog (Fragment).
Length = 244
Score = 260 bits (665), Expect = 3e-68
Identities = 131/224 (58%), Positives = 179/224 (79%)
Frame = +3
Query: 249 GKGKTEIKRIENTTNRQVTFCKRRNGLLKKAYELSVLCDAEVALIVFSSRGRLYEYANNS 428
G+GK EIKRIENTTNRQVTFCKRRNGLLKKAYELSVLCDAE+ALIVFSSRGRLYEYANNS
Sbjct: 21 GRGKIEIKRIENTTNRQVTFCKRRNGLLKKAYELSVLCDAEIALIVFSSRGRLYEYANNS 80
Query: 429 VKGTIERYKKATSDNSSAAGTIAEVTIQHYKQESARLRQQIVNLQNSNRALIGDSITTMS 608
VK TIERYKKA+ ++S G+++E Q Y+QES++LR+QI ++QN NR ++G+++++++
Sbjct: 81 VKSTIERYKKAS--DTSNPGSVSETNAQFYQQESSKLRRQIRDIQNLNRHIMGEALSSLT 138
Query: 609 HKELKHLETRLDKALGKIRAKKNDVLCSEVEYMQRREMELQNDNLYLRSRVDENERAQQT 788
+ELK+LE RL+K + +IR+KKN++L +E+EYMQ+RE+ELQN N+YLR+++ ENER QQ
Sbjct: 139 FRELKNLEGRLEKGISRIRSKKNELLFAEIEYMQKREIELQNANMYLRAKIAENERNQQQ 198
Query: 789 ANMMGAPSTSEYQQHGFTPYDPIRSFLQFNIVQQPQFYSQQEDR 920
+M P + PYD RSFL N+++ P + ++D+
Sbjct: 199 TELM--PGSVYETMPSSQPYD--RSFLVANLLEPPNHHYSRQDQ 238
>sptr|Q40900|Q40900 Agamous protein.
Length = 247
Score = 260 bits (665), Expect = 3e-68
Identities = 132/226 (58%), Positives = 183/226 (80%), Gaps = 4/226 (1%)
Frame = +3
Query: 249 GKGKTEIKRIENTTNRQVTFCKRRNGLLKKAYELSVLCDAEVALIVFSSRGRLYEYANNS 428
G+GK EIKRIENTTNRQVTFCKRRNGLLKKAYELSVLCDAEVALIVFSSRGRLYEYANNS
Sbjct: 18 GRGKIEIKRIENTTNRQVTFCKRRNGLLKKAYELSVLCDAEVALIVFSSRGRLYEYANNS 77
Query: 429 VKGTIERYKKATSDNSSAAGTIAEVTIQHYKQESARLRQQIVNLQNSNRALIGDSITTMS 608
V+ TI+RYKK +D S++ G+++E Q+Y+QE+A+LR+QI ++Q NR ++G++++++S
Sbjct: 78 VRATIDRYKKHHAD-STSTGSVSEANTQYYQQEAAKLRRQIRDIQTYNRQIVGEALSSLS 136
Query: 609 HKELKHLETRLDKALGKIRAKKNDVLCSEVEYMQRREMELQNDNLYLRSRVDENERAQQT 788
++LK+LE +L+KA+G++R+KKN++L SE+E MQ+RE+E+QN N+YLR+++ E ERA Q
Sbjct: 137 PRDLKNLEGKLEKAIGRVRSKKNELLFSEIELMQKREIEMQNANMYLRAKIAEVERATQQ 196
Query: 789 ANMMGAPSTSEYQQHGFT----PYDPIRSFLQFNIVQQPQFYSQQE 914
N+M SEYQQ + PYD R+FL N+++ YS+Q+
Sbjct: 197 MNLMPG-GGSEYQQQPMSSTSQPYD-ARNFLPVNLLEPNPHYSRQD 240
>sptr|Q8RVK1|Q8RVK1 AP3-like protein.
Length = 244
Score = 260 bits (664), Expect = 4e-68
Identities = 133/226 (58%), Positives = 176/226 (77%)
Frame = +3
Query: 249 GKGKTEIKRIENTTNRQVTFCKRRNGLLKKAYELSVLCDAEVALIVFSSRGRLYEYANNS 428
G+GK EIKRIENTTNRQVTFCKRRNGLLKKAYELSVLCDAEVAL+ FSSRGRLYEYANNS
Sbjct: 17 GRGKIEIKRIENTTNRQVTFCKRRNGLLKKAYELSVLCDAEVALVAFSSRGRLYEYANNS 76
Query: 429 VKGTIERYKKATSDNSSAAGTIAEVTIQHYKQESARLRQQIVNLQNSNRALIGDSITTMS 608
VK TIERYKKA+ +SS G++AEV Q Y+QE+ +LR QI NLQN+NR ++G+SI +
Sbjct: 77 VKATIERYKKAS--DSSNTGSVAEVNAQFYQQEADKLRNQIRNLQNANRHMLGESIGGLP 134
Query: 609 HKELKHLETRLDKALGKIRAKKNDVLCSEVEYMQRREMELQNDNLYLRSRVDENERAQQT 788
KELK LE+RL+K + +IR+KKN++L +E+EYMQ+RE++L N+N LR+++ ENER QQ+
Sbjct: 135 MKELKSLESRLEKGISRIRSKKNELLFAEIEYMQKREIDLHNNNQLLRAKIAENERKQQS 194
Query: 789 ANMMGAPSTSEYQQHGFTPYDPIRSFLQFNIVQQPQFYSQQEDRKD 926
N+M S++ ++ PYD R++ Q + +Q Y + ++D
Sbjct: 195 MNLMPGGSSANFEALHSQPYDS-RNYFQVDALQPATNYYNPQLQQD 239
>sptr|Q08711|Q08711 Fbp6 protein.
Length = 247
Score = 259 bits (661), Expect = 8e-68
Identities = 132/226 (58%), Positives = 182/226 (80%), Gaps = 4/226 (1%)
Frame = +3
Query: 249 GKGKTEIKRIENTTNRQVTFCKRRNGLLKKAYELSVLCDAEVALIVFSSRGRLYEYANNS 428
G+GK EIKRIENTTNRQVTFCKRRNGLLKKAYELSVLCDAEVALIVFSSRGRLYEYANNS
Sbjct: 18 GRGKIEIKRIENTTNRQVTFCKRRNGLLKKAYELSVLCDAEVALIVFSSRGRLYEYANNS 77
Query: 429 VKGTIERYKKATSDNSSAAGTIAEVTIQHYKQESARLRQQIVNLQNSNRALIGDSITTMS 608
V+ TI+RYKK +D S++ G+++E Q+Y+QE+A+LR+QI ++Q NR ++G++++++S
Sbjct: 78 VRATIDRYKKHHAD-STSTGSVSEANTQYYQQEAAKLRRQIRDIQTYNRQIVGEALSSLS 136
Query: 609 HKELKHLETRLDKALGKIRAKKNDVLCSEVEYMQRREMELQNDNLYLRSRVDENERAQQT 788
+ LK+LE +L+KA+G++R+KKN++L SE+E MQ+RE+E+QN N+YLR+++ E ERA Q
Sbjct: 137 PRGLKNLEGKLEKAIGRVRSKKNELLFSEIELMQKREIEMQNANMYLRAKIAEVERATQQ 196
Query: 789 ANMMGAPSTSEYQQHGFT----PYDPIRSFLQFNIVQQPQFYSQQE 914
N+M SEYQQ + PYD R+FL N+++ YS+Q+
Sbjct: 197 MNLMHG-GGSEYQQQPMSSTSQPYD-ARNFLPVNLLEPNPHYSRQD 240
>sw|Q40872|AG_PANGI Floral homeotic protein AGAMOUS (GAG2).
Length = 242
Score = 258 bits (659), Expect = 1e-67
Identities = 128/222 (57%), Positives = 182/222 (81%)
Frame = +3
Query: 249 GKGKTEIKRIENTTNRQVTFCKRRNGLLKKAYELSVLCDAEVALIVFSSRGRLYEYANNS 428
G+GK EIKRIENTTNRQVTFCKRRNGLLKKAYELSVLCDAEVALIVFS+RGRLYEYANNS
Sbjct: 18 GRGKIEIKRIENTTNRQVTFCKRRNGLLKKAYELSVLCDAEVALIVFSTRGRLYEYANNS 77
Query: 429 VKGTIERYKKATSDNSSAAGTIAEVTIQHYKQESARLRQQIVNLQNSNRALIGDSITTMS 608
VKGTIERYKKA +D+ + + +++E Q Y+QE+++LRQ+I ++Q +NR ++G+S+ +++
Sbjct: 78 VKGTIERYKKACTDSPNTS-SVSEANAQFYQQEASKLRQEISSIQKNNRNMMGESLGSLT 136
Query: 609 HKELKHLETRLDKALGKIRAKKNDVLCSEVEYMQRREMELQNDNLYLRSRVDENERAQQT 788
++LK LET+L+K + +IR+KKN++L +E+EYMQ++E++L N+N YLR+++ ENERAQQ
Sbjct: 137 VRDLKGLETKLEKGISRIRSKKNELLFAEIEYMQKKEIDLHNNNQYLRAKIAENERAQQH 196
Query: 789 ANMMGAPSTSEYQQHGFTPYDPIRSFLQFNIVQQPQFYSQQE 914
N+M P +S+Y+ +D R+++Q N +Q YS+Q+
Sbjct: 197 MNLM--PGSSDYELAPPQSFDG-RNYIQLNGLQPNNHYSRQD 235
>sw|Q43585|AG_TOBAC Floral homeotic protein AGAMOUS (NAG1).
Length = 248
Score = 258 bits (659), Expect = 1e-67
Identities = 134/225 (59%), Positives = 174/225 (77%), Gaps = 3/225 (1%)
Frame = +3
Query: 249 GKGKTEIKRIENTTNRQVTFCKRRNGLLKKAYELSVLCDAEVALIVFSSRGRLYEYANNS 428
G+GK EIKRIENTTNRQVTFCKRRNGLLKKAYELSVLCDAEVALIVFSSRGRLYEYANNS
Sbjct: 18 GRGKIEIKRIENTTNRQVTFCKRRNGLLKKAYELSVLCDAEVALIVFSSRGRLYEYANNS 77
Query: 429 VKGTIERYKKATSDNSSAAGTIAEVTIQHYKQESARLRQQIVNLQNSNRALIGDSITTMS 608
VK TIERYKKA SD SS G+I+E Q+Y+QE+++LR QI NLQN NR ++G+S+ +S
Sbjct: 78 VKATIERYKKACSD-SSNTGSISEANAQYYQQEASKLRAQIGNLQNQNRNMLGESLAALS 136
Query: 609 HKELKHLETRLDKALGKIRAKKNDVLCSEVEYMQRREMELQNDNLYLRSRVDENERA--- 779
++LK+LE +++K + KIR+KKN++L +E+EYMQ+RE++L N+N YLR+++ E ERA
Sbjct: 137 LRDLKNLEQKIEKGISKIRSKKNELLFAEIEYMQKREIDLHNNNQYLRAKIAETERAQQQ 196
Query: 780 QQTANMMGAPSTSEYQQHGFTPYDPIRSFLQFNIVQQPQFYSQQE 914
QQ M P +S Y+ R++LQ N +Q Y++Q+
Sbjct: 197 QQQQQMNLMPGSSSYELVPPPHQFDTRNYLQVNGLQTNNHYTRQD 241
>sptr|Q9SBK1|Q9SBK1 Agamous-like putative transcription factor.
Length = 237
Score = 257 bits (657), Expect = 2e-67
Identities = 135/223 (60%), Positives = 176/223 (78%)
Frame = +3
Query: 249 GKGKTEIKRIENTTNRQVTFCKRRNGLLKKAYELSVLCDAEVALIVFSSRGRLYEYANNS 428
G+GK EIKRIENTTNRQVTFCKRRNGLLKKAYELSVLCDAEVALIVFSSRGRLYEYANNS
Sbjct: 18 GRGKIEIKRIENTTNRQVTFCKRRNGLLKKAYELSVLCDAEVALIVFSSRGRLYEYANNS 77
Query: 429 VKGTIERYKKATSDNSSAAGTIAEVTIQHYKQESARLRQQIVNLQNSNRALIGDSITTMS 608
VK TI+RYKKA+SD SS G+ +E Q Y+QE+A+LR QI NLQNSNR ++G+S+++++
Sbjct: 78 VKATIDRYKKASSD-SSNTGSTSEANTQFYQQEAAKLRVQIGNLQNSNRNMLGESLSSLT 136
Query: 609 HKELKHLETRLDKALGKIRAKKNDVLCSEVEYMQRREMELQNDNLYLRSRVDENERAQQT 788
K+LK LET+L+K + +IR+KKN++L +E+EYM++RE++L N+N LR+++ E+ER
Sbjct: 137 AKDLKGLETKLEKGISRIRSKKNELLFAEIEYMRKREIDLHNNNQMLRAKIAESER---N 193
Query: 789 ANMMGAPSTSEYQQHGFTPYDPIRSFLQFNIVQQPQFYSQQED 917
NMMG E++ PYDP R F Q N +Q Y +Q++
Sbjct: 194 VNMMG----GEFELMQSHPYDP-RDFFQVNGLQHNHQYPRQDN 231
>sptr|O64958|O64958 CUM1.
Length = 262
Score = 257 bits (657), Expect = 2e-67
Identities = 135/223 (60%), Positives = 176/223 (78%)
Frame = +3
Query: 249 GKGKTEIKRIENTTNRQVTFCKRRNGLLKKAYELSVLCDAEVALIVFSSRGRLYEYANNS 428
G+GK EIKRIENTTNRQVTFCKRRNGLLKKAYELSVLCDAEVALIVFSSRGRLYEYANNS
Sbjct: 43 GRGKIEIKRIENTTNRQVTFCKRRNGLLKKAYELSVLCDAEVALIVFSSRGRLYEYANNS 102
Query: 429 VKGTIERYKKATSDNSSAAGTIAEVTIQHYKQESARLRQQIVNLQNSNRALIGDSITTMS 608
VK TI+RYKKA+SD SS G+ +E Q Y+QE+A+LR QI NLQNSNR ++G+S+++++
Sbjct: 103 VKATIDRYKKASSD-SSNTGSTSEANTQFYQQEAAKLRVQIGNLQNSNRNMLGESLSSLT 161
Query: 609 HKELKHLETRLDKALGKIRAKKNDVLCSEVEYMQRREMELQNDNLYLRSRVDENERAQQT 788
K+LK LET+L+K + +IR+KKN++L +E+EYM++RE++L N+N LR+++ E+ER
Sbjct: 162 AKDLKGLETKLEKGISRIRSKKNELLFAEIEYMRKREIDLHNNNQMLRAKIAESER---N 218
Query: 789 ANMMGAPSTSEYQQHGFTPYDPIRSFLQFNIVQQPQFYSQQED 917
NMMG E++ PYDP R F Q N +Q Y +Q++
Sbjct: 219 VNMMG----GEFELMQSHPYDP-RDFFQVNGLQHNHQYPRQDN 256
>sptr|Q84LC4|Q84LC4 MADS-box transcriptional factor HAM45.
Length = 267
Score = 257 bits (656), Expect = 3e-67
Identities = 142/231 (61%), Positives = 174/231 (75%), Gaps = 9/231 (3%)
Frame = +3
Query: 249 GKGKTEIKRIENTTNRQVTFCKRRNGLLKKAYELSVLCDAEVALIVFSSRGRLYEYANNS 428
GKGK EIKRIENTTNRQVTFCKRRNGLLKKAYELSVLCDAEVALIVFSSRGRLYEYANNS
Sbjct: 37 GKGKIEIKRIENTTNRQVTFCKRRNGLLKKAYELSVLCDAEVALIVFSSRGRLYEYANNS 96
Query: 429 VKGTIERYKKATSDNSSAAGTIAEVTIQHYKQESARLRQQIVNLQNSNRA----LIGDSI 596
V+GTI+RYKK+ D S G++AE Q Y+QE+ +LRQQI NLQN NR ++G+S+
Sbjct: 97 VRGTIDRYKKSCLDPPS-TGSVAEANAQFYQQEATKLRQQIANLQNQNRQFYRNIMGESL 155
Query: 597 TTMSHKELKHLETRLDKALGKIRAKKNDVLCSEVEYMQRREMELQNDNLYLRSRVDENER 776
M KELK+LE++L+KA+ +IRAKKN++L +E+EYMQ+RE+EL N N +LR+R+ ENER
Sbjct: 156 ADMPGKELKNLESKLEKAINRIRAKKNELLFAEIEYMQKRELELHNSNQFLRARISENER 215
Query: 777 AQQTANMMGAPSTSEYQQHGFTPYDPIRSF-----LQFNIVQQPQFYSQQE 914
AQQ +M P +S Y G P +SF LQ N +Q YS Q+
Sbjct: 216 AQQ-QHMSLMPGSSGYNDLG-----PHQSFDGLNDLQTNELQLNNNYSCQD 260
>sw|Q40885|AG_PETHY Floral homeotic protein AGAMOUS (pMADS3).
Length = 242
Score = 256 bits (655), Expect = 4e-67
Identities = 134/222 (60%), Positives = 173/222 (77%)
Frame = +3
Query: 249 GKGKTEIKRIENTTNRQVTFCKRRNGLLKKAYELSVLCDAEVALIVFSSRGRLYEYANNS 428
G+GK EIKRIENTTNRQVTFCKRRNGLLKKAYELSVLCDAEVALIVFSSRGRLYEYANNS
Sbjct: 18 GRGKIEIKRIENTTNRQVTFCKRRNGLLKKAYELSVLCDAEVALIVFSSRGRLYEYANNS 77
Query: 429 VKGTIERYKKATSDNSSAAGTIAEVTIQHYKQESARLRQQIVNLQNSNRALIGDSITTMS 608
VK TIERYKKA SD SS G+IAE Q+Y+QE+++LR QI NLQN NR +G+S+ ++
Sbjct: 78 VKATIERYKKACSD-SSNTGSIAEANAQYYQQEASKLRAQIGNLQNQNRNFLGESLAALN 136
Query: 609 HKELKHLETRLDKALGKIRAKKNDVLCSEVEYMQRREMELQNDNLYLRSRVDENERAQQT 788
++L++LE +++K + KIRAKKN++L +E+EYMQ+RE++L N+N YLR+++ E ER+QQ
Sbjct: 137 LRDLRNLEQKIEKGISKIRAKKNELLFAEIEYMQKREIDLHNNNQYLRAKIAETERSQQ- 195
Query: 789 ANMMGAPSTSEYQQHGFTPYDPIRSFLQFNIVQQPQFYSQQE 914
N+M P +S Y R++LQ N +Q Y +Q+
Sbjct: 196 MNLM--PGSSSYDLVPPQQSFDARNYLQVNGLQTNNHYPRQD 235
Database: /db/trembl-ebi/tmp/swall
Posted date: Sep 12, 2003 8:28 PM
Number of letters in database: 397,940,306
Number of sequences in database: 1,242,019
Lambda K H
0.318 0.135 0.401
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 855,016,554
Number of Sequences: 1242019
Number of extensions: 17111604
Number of successful extensions: 50353
Number of sequences better than 10.0: 30
Number of HSP's better than 10.0 without gapping: 47526
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 49770
length of database: 397,940,306
effective HSP length: 126
effective length of database: 241,445,912
effective search space used: 78228475488
frameshift window, decay const: 50, 0.1
T: 12
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)

