BLASTX 2.2.6 [Apr-09-2003]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
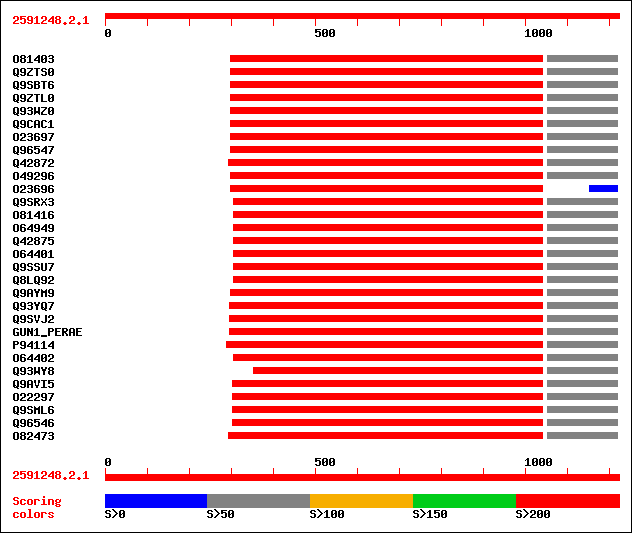
Query= 2591248.2.1
(1222 letters)
Database: /db/uniprot/tmp/swall
1,395,590 sequences; 442,889,342 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
sptr|O81403|O81403 Endo-1,4-beta-glucanase precursor (EC 3.2.1.4). 395 e-127
sptr|Q9ZTS0|Q9ZTS0 Putative cellulase. 394 e-127
sptr|Q9SBT6|Q9SBT6 Endo-beta-1,4-glucanase (EC 3.2.1.4). 393 e-127
sptr|Q9ZTL0|Q9ZTL0 Cellulase. 389 e-126
sptr|Q93WZ0|Q93WZ0 Endo-beta-1,4-glucanase precursor (EC 3.2.1.4). 385 e-125
sptr|Q9CAC1|Q9CAC1 Endo-1,4-beta-glucanase (At1g70710/F5A18_11). 384 e-125
sptr|O23697|O23697 Endo-1,4-beta-glucanase. 384 e-124
sptr|Q96547|Q96547 Cellulase (EC 3.2.1.4). 375 e-123
sptr|Q42872|Q42872 Endo-1,4-beta-glucanase precursor (EC 3.2.1.4). 376 e-123
sptr|O49296|O49296 T26J12.2 protein (F26F24.6). 368 e-119
sptr|O23696|O23696 Endo-1,4-beta-glucanase. 382 e-108
sptr|Q9SRX3|Q9SRX3 Endo-1,4-beta glucanase. 288 2e-96
sptr|O81416|O81416 T2H3.5 protein (Putative endo-1, 4-beta gluca... 291 3e-96
sptr|O64949|O64949 Endo-1,4-beta glucanase. 288 3e-96
sptr|Q42875|Q42875 Endo-1,4-beta-glucanase precursor (EC 3.2.1.4). 293 6e-95
sptr|O64401|O64401 Endo-beta-1,4-glucanase. 285 2e-94
sptr|Q9SSU7|Q9SSU7 Endo-1,4-beta-glucanase. 290 4e-93
sptr|Q8LQ92|Q8LQ92 Putative endo-1,3(4)-beta-glucanase. 282 8e-93
sptr|Q9AYM9|Q9AYM9 Beta-1,4-glucanase. 273 5e-92
sptr|Q93YQ7|Q93YQ7 Putative endo-1, 4-beta-glucanase. 287 2e-91
sptr|Q9SVJ2|Q9SVJ2 Putative endo-1, 4-beta-glucanase. 287 2e-91
sw|P05522|GUN1_PERAE Endoglucanase 1 precursor (EC 3.2.1.4) (End... 275 3e-90
sptr|P94114|P94114 Endo-beta-1,4-glucanase (EC 3.2.1.4) (Cellula... 273 3e-90
sptr|O64402|O64402 Endo-beta-1,4-glucanase. 270 1e-88
sptr|Q93WY8|Q93WY8 Endo-beta-1,4-glucanase (EC 3.2.1.4) (Fragment). 270 3e-88
sptr|Q9AVI5|Q9AVI5 Endo-1,4-beta glucanase precursor. 263 5e-88
sptr|O22297|O22297 Acidic cellulase. 262 1e-87
sptr|Q9SML6|Q9SML6 Endo-beta-1,4-glucanase precursor (EC 3.2.1.4). 264 2e-87
sptr|Q96546|Q96546 Endo-beta-1,4-glucanase (EC 3.2.1.4). 264 3e-87
sptr|O82473|O82473 Endo-1,4-beta-glucanase (EC 3.2.1.4). 267 5e-87
>sptr|O81403|O81403 Endo-1,4-beta-glucanase precursor (EC 3.2.1.4).
Length = 496
Score = 395 bits (1014), Expect(2) = e-127
Identities = 189/247 (76%), Positives = 216/247 (87%)
Frame = -1
Query: 1039 AWLHKATRRREYREYIKRNEVVLGASDAINEFGWDNKHAGINVLISKEVLMGKDEYFQSF 860
AWLHKA+RRR+YREYI RNEVVL A D INEFGWDNKHAGIN+LISKEVLMGK +YF+SF
Sbjct: 251 AWLHKASRRRQYREYIVRNEVVLRAGDTINEFGWDNKHAGINILISKEVLMGKADYFESF 310
Query: 859 RVNADNFMCTLLPGISNHPQIQYSPGGLLFKVGSSNMQHVTQLSFLLLAYSNYLSHAGGR 680
+ NAD F+C++LPG++ H Q+QYSPGGL+FK G SNMQHVT LSFLLL YSNYLSHA
Sbjct: 311 KQNADGFICSVLPGLA-HTQVQYSPGGLIFKPGGSNMQHVTSLSFLLLTYSNYLSHANKN 369
Query: 679 VSCGSSSASPVQLRRVAKRQVDYILGDNPLRMSYMVGYGARFPRRIHHRASSLPSVAAHP 500
V CG +SASP L+++AKRQVDYILGDNPLRMSYMVGYG R+P+RIHHR SSLPSV AHP
Sbjct: 370 VPCGMTSASPAFLKQLAKRQVDYILGDNPLRMSYMVGYGPRYPQRIHHRGSSLPSVQAHP 429
Query: 499 ARIGCKAGAAYYTSPAPNPNLLVGAVVGGPTDDSDAFPDARAVFQQSEPTTYINAPLMGL 320
ARIGCKAG+ Y+ SP PNPN LVGAVVGGP + SDAFPD+R FQ+SEPTTYINAPL+GL
Sbjct: 430 ARIGCKAGSRYFMSPNPNPNKLVGAVVGGP-NSSDAFPDSRPYFQESEPTTYINAPLVGL 488
Query: 319 LAYFSAH 299
L+YF+AH
Sbjct: 489 LSYFAAH 495
Score = 84.7 bits (208), Expect(2) = e-127
Identities = 41/56 (73%), Positives = 46/56 (82%)
Frame = -2
Query: 1218 SIVFRDADPAYSKRLLDRAVAVFEFADKYRGPYSSSLHAAVCPCYCDYSGYQDELL 1051
SIVFR DPAYS+ LL+RAV VFEFAD +RG YSSSL AVCP YCD +G+QDELL
Sbjct: 192 SIVFRSHDPAYSRLLLNRAVRVFEFADTHRGGYSSSLKNAVCPFYCDVNGFQDELL 247
>sptr|Q9ZTS0|Q9ZTS0 Putative cellulase.
Length = 496
Score = 394 bits (1012), Expect(2) = e-127
Identities = 188/247 (76%), Positives = 216/247 (87%)
Frame = -1
Query: 1039 AWLHKATRRREYREYIKRNEVVLGASDAINEFGWDNKHAGINVLISKEVLMGKDEYFQSF 860
AWLHKA+RRR+YREYI RNEV+L A D INEFGWDNKHAGIN+LISKEVLMGK +YF+SF
Sbjct: 251 AWLHKASRRRQYREYIVRNEVILRAGDTINEFGWDNKHAGINILISKEVLMGKADYFESF 310
Query: 859 RVNADNFMCTLLPGISNHPQIQYSPGGLLFKVGSSNMQHVTQLSFLLLAYSNYLSHAGGR 680
+ NAD F+C++LPG++ H Q+QYSPGGL+FK G SNMQHVT LSFLLL YSNYLSHA
Sbjct: 311 KQNADGFICSVLPGLA-HTQVQYSPGGLIFKPGGSNMQHVTSLSFLLLTYSNYLSHANKN 369
Query: 679 VSCGSSSASPVQLRRVAKRQVDYILGDNPLRMSYMVGYGARFPRRIHHRASSLPSVAAHP 500
V CG +SASP L+++AKRQVDYILGDNPLRMSYMVGYG R+P+RIHHR SSLPSV AHP
Sbjct: 370 VPCGMTSASPAFLKQLAKRQVDYILGDNPLRMSYMVGYGPRYPQRIHHRGSSLPSVQAHP 429
Query: 499 ARIGCKAGAAYYTSPAPNPNLLVGAVVGGPTDDSDAFPDARAVFQQSEPTTYINAPLMGL 320
ARIGCKAG+ Y+ SP PNPN LVGAVVGGP + SDAFPD+R FQ+SEPTTYINAPL+GL
Sbjct: 430 ARIGCKAGSHYFLSPNPNPNKLVGAVVGGP-NSSDAFPDSRPYFQESEPTTYINAPLVGL 488
Query: 319 LAYFSAH 299
L+YF+AH
Sbjct: 489 LSYFAAH 495
Score = 85.5 bits (210), Expect(2) = e-127
Identities = 41/56 (73%), Positives = 46/56 (82%)
Frame = -2
Query: 1218 SIVFRDADPAYSKRLLDRAVAVFEFADKYRGPYSSSLHAAVCPCYCDYSGYQDELL 1051
SIVFR DPAYS+ LL+RAV VFEFAD +RG YSSSL AVCP YCD +G+QDELL
Sbjct: 192 SIVFRSRDPAYSRLLLNRAVKVFEFADTHRGAYSSSLKNAVCPFYCDVNGFQDELL 247
>sptr|Q9SBT6|Q9SBT6 Endo-beta-1,4-glucanase (EC 3.2.1.4).
Length = 496
Score = 393 bits (1009), Expect(2) = e-127
Identities = 188/247 (76%), Positives = 215/247 (87%)
Frame = -1
Query: 1039 AWLHKATRRREYREYIKRNEVVLGASDAINEFGWDNKHAGINVLISKEVLMGKDEYFQSF 860
AWLHKA+RRR+YREYI RNEV+L A D INEFGWDNKHAGIN+LISKEVLMGK +YF+SF
Sbjct: 251 AWLHKASRRRQYREYIVRNEVILRAGDTINEFGWDNKHAGINILISKEVLMGKADYFESF 310
Query: 859 RVNADNFMCTLLPGISNHPQIQYSPGGLLFKVGSSNMQHVTQLSFLLLAYSNYLSHAGGR 680
+ NAD F+C++LPG++ H Q QYSPGGL+FK G SNMQHVT LSFLLL YSNYLSHA
Sbjct: 311 KQNADGFICSVLPGLA-HTQAQYSPGGLIFKPGGSNMQHVTSLSFLLLTYSNYLSHANKN 369
Query: 679 VSCGSSSASPVQLRRVAKRQVDYILGDNPLRMSYMVGYGARFPRRIHHRASSLPSVAAHP 500
V CG +SASP L+++AKRQVDYILGDNPLRMSYMVGYG R+P+RIHHR SSLPSV AHP
Sbjct: 370 VPCGMTSASPAFLKQLAKRQVDYILGDNPLRMSYMVGYGPRYPQRIHHRGSSLPSVQAHP 429
Query: 499 ARIGCKAGAAYYTSPAPNPNLLVGAVVGGPTDDSDAFPDARAVFQQSEPTTYINAPLMGL 320
ARIGCKAG+ Y+ SP PNPN LVGAVVGGP + SDAFPD+R FQ+SEPTTYINAPL+GL
Sbjct: 430 ARIGCKAGSRYFLSPNPNPNKLVGAVVGGP-NSSDAFPDSRPYFQESEPTTYINAPLVGL 488
Query: 319 LAYFSAH 299
L+YF+AH
Sbjct: 489 LSYFAAH 495
Score = 85.5 bits (210), Expect(2) = e-127
Identities = 41/56 (73%), Positives = 46/56 (82%)
Frame = -2
Query: 1218 SIVFRDADPAYSKRLLDRAVAVFEFADKYRGPYSSSLHAAVCPCYCDYSGYQDELL 1051
SIVFR DPAYS+ LL+RAV VFEFAD +RG YSSSL AVCP YCD +G+QDELL
Sbjct: 192 SIVFRSRDPAYSRLLLNRAVKVFEFADTHRGAYSSSLKNAVCPFYCDVNGFQDELL 247
>sptr|Q9ZTL0|Q9ZTL0 Cellulase.
Length = 496
Score = 389 bits (1000), Expect(2) = e-126
Identities = 186/247 (75%), Positives = 213/247 (86%)
Frame = -1
Query: 1039 AWLHKATRRREYREYIKRNEVVLGASDAINEFGWDNKHAGINVLISKEVLMGKDEYFQSF 860
AWLHKA+RRR+YREYI RNEV+L A D INEFGWDNKHAGIN+LISKEVLMGK +YF+SF
Sbjct: 251 AWLHKASRRRQYREYIVRNEVILRAGDTINEFGWDNKHAGINILISKEVLMGKSDYFESF 310
Query: 859 RVNADNFMCTLLPGISNHPQIQYSPGGLLFKVGSSNMQHVTQLSFLLLAYSNYLSHAGGR 680
+ NAD F+C++LPG++ H Q+QYSPGGL+FK G SNMQHVT LSFLLL YSNYLSHA
Sbjct: 311 KQNADGFICSVLPGLA-HTQVQYSPGGLIFKPGGSNMQHVTSLSFLLLTYSNYLSHANKN 369
Query: 679 VSCGSSSASPVQLRRVAKRQVDYILGDNPLRMSYMVGYGARFPRRIHHRASSLPSVAAHP 500
V CG +SAS ++AKRQVDYILGDNPLRMSYMVGYG R+P+RIHHR SSLPSV AHP
Sbjct: 370 VPCGMTSASRPSSNKLAKRQVDYILGDNPLRMSYMVGYGPRYPQRIHHRGSSLPSVQAHP 429
Query: 499 ARIGCKAGAAYYTSPAPNPNLLVGAVVGGPTDDSDAFPDARAVFQQSEPTTYINAPLMGL 320
ARIGCKAG+ Y+ SP PNPN LVGAVVGGP + SDAFPD+R FQ+SEPTTYINAPL+GL
Sbjct: 430 ARIGCKAGSRYFLSPNPNPNKLVGAVVGGP-NSSDAFPDSRPYFQESEPTTYINAPLVGL 488
Query: 319 LAYFSAH 299
L+YF+AH
Sbjct: 489 LSYFAAH 495
Score = 85.5 bits (210), Expect(2) = e-126
Identities = 41/56 (73%), Positives = 46/56 (82%)
Frame = -2
Query: 1218 SIVFRDADPAYSKRLLDRAVAVFEFADKYRGPYSSSLHAAVCPCYCDYSGYQDELL 1051
SIVFR DPAYS+ LL+RAV VFEFAD +RG YSSSL AVCP YCD +G+QDELL
Sbjct: 192 SIVFRSRDPAYSRLLLNRAVKVFEFADTHRGAYSSSLKNAVCPFYCDVNGFQDELL 247
>sptr|Q93WZ0|Q93WZ0 Endo-beta-1,4-glucanase precursor (EC 3.2.1.4).
Length = 500
Score = 385 bits (989), Expect(2) = e-125
Identities = 188/247 (76%), Positives = 211/247 (85%)
Frame = -1
Query: 1039 AWLHKATRRREYREYIKRNEVVLGASDAINEFGWDNKHAGINVLISKEVLMGKDEYFQSF 860
AWLHKATRRR+YREYI +NEVVL A D INEFGWDNKHAGINVLISKEVLMG+ +SF
Sbjct: 255 AWLHKATRRRQYREYIVKNEVVLRAGDTINEFGWDNKHAGINVLISKEVLMGRAPDLKSF 314
Query: 859 RVNADNFMCTLLPGISNHPQIQYSPGGLLFKVGSSNMQHVTQLSFLLLAYSNYLSHAGGR 680
+VNAD F+C++LPGIS HPQ+QYSPGGL+ K G NMQHVT LSFLLLAYSNYLSHA
Sbjct: 315 QVNADAFICSILPGIS-HPQVQYSPGGLIVKPGVCNMQHVTSLSFLLLAYSNYLSHANHA 373
Query: 679 VSCGSSSASPVQLRRVAKRQVDYILGDNPLRMSYMVGYGARFPRRIHHRASSLPSVAAHP 500
V CGS SA+P L+ +AKRQVDYILGDNP RMSYMVGYG R+P RIHHR SSLPSVAAHP
Sbjct: 374 VPCGSISATPALLKHIAKRQVDYILGDNPQRMSYMVGYGPRYPLRIHHRGSSLPSVAAHP 433
Query: 499 ARIGCKAGAAYYTSPAPNPNLLVGAVVGGPTDDSDAFPDARAVFQQSEPTTYINAPLMGL 320
ARIGCK G+ Y+ SP PNPN L+GAVVGGP + +D+FPDAR FQ+SEPTTY+NAPL+GL
Sbjct: 434 ARIGCKGGSNYFLSPNPNPNRLIGAVVGGP-NITDSFPDARPFFQESEPTTYVNAPLVGL 492
Query: 319 LAYFSAH 299
LAYFSAH
Sbjct: 493 LAYFSAH 499
Score = 88.2 bits (217), Expect(2) = e-125
Identities = 41/56 (73%), Positives = 47/56 (83%)
Frame = -2
Query: 1218 SIVFRDADPAYSKRLLDRAVAVFEFADKYRGPYSSSLHAAVCPCYCDYSGYQDELL 1051
SIVFR D +YS LLDRAV VFEFA+++RG YSSSLH+AVCP YCD+ GYQDELL
Sbjct: 196 SIVFRSLDSSYSGLLLDRAVKVFEFANRHRGAYSSSLHSAVCPFYCDFDGYQDELL 251
>sptr|Q9CAC1|Q9CAC1 Endo-1,4-beta-glucanase (At1g70710/F5A18_11).
Length = 492
Score = 384 bits (987), Expect(2) = e-125
Identities = 186/247 (75%), Positives = 212/247 (85%)
Frame = -1
Query: 1039 AWLHKATRRREYREYIKRNEVVLGASDAINEFGWDNKHAGINVLISKEVLMGKDEYFQSF 860
AWLHKA+R+R YRE+I +NEV+L A D INEFGWDNKHAGINVLISKEVLMGK EYF+SF
Sbjct: 244 AWLHKASRKRAYREFIVKNEVILKAGDTINEFGWDNKHAGINVLISKEVLMGKAEYFESF 303
Query: 859 RVNADNFMCTLLPGISNHPQIQYSPGGLLFKVGSSNMQHVTQLSFLLLAYSNYLSHAGGR 680
+ NAD F+C++LPGIS HPQ+QYS GGLL K G SNMQHVT LSFLLLAYSNYLSHA
Sbjct: 304 KQNADGFICSILPGIS-HPQVQYSRGGLLVKTGGSNMQHVTSLSFLLLAYSNYLSHAKKV 362
Query: 679 VSCGSSSASPVQLRRVAKRQVDYILGDNPLRMSYMVGYGARFPRRIHHRASSLPSVAAHP 500
V CG +ASP LR++AKRQVDYILGDNP+ +SYMVGYG +FPRRIHHR SS+PSV+AHP
Sbjct: 363 VPCGELTASPSLLRQIAKRQVDYILGDNPMGLSYMVGYGQKFPRRIHHRGSSVPSVSAHP 422
Query: 499 ARIGCKAGAAYYTSPAPNPNLLVGAVVGGPTDDSDAFPDARAVFQQSEPTTYINAPLMGL 320
+ IGCK G+ Y+ SP PNPNLLVGAVVGGP + +DAFPD+R FQQSEPTTYINAPL+GL
Sbjct: 423 SHIGCKEGSRYFLSPNPNPNLLVGAVVGGP-NVTDAFPDSRPYFQQSEPTTYINAPLVGL 481
Query: 319 LAYFSAH 299
L YFSAH
Sbjct: 482 LGYFSAH 488
Score = 87.4 bits (215), Expect(2) = e-125
Identities = 40/56 (71%), Positives = 47/56 (83%)
Frame = -2
Query: 1218 SIVFRDADPAYSKRLLDRAVAVFEFADKYRGPYSSSLHAAVCPCYCDYSGYQDELL 1051
SIVFR DPAYS+ LLDRA VF FA++YRG YS+SL+ AVCP YCD++GYQDELL
Sbjct: 185 SIVFRKRDPAYSRLLLDRATRVFAFANRYRGAYSNSLYHAVCPFYCDFNGYQDELL 240
>sptr|O23697|O23697 Endo-1,4-beta-glucanase.
Length = 492
Score = 384 bits (987), Expect(2) = e-124
Identities = 186/247 (75%), Positives = 212/247 (85%)
Frame = -1
Query: 1039 AWLHKATRRREYREYIKRNEVVLGASDAINEFGWDNKHAGINVLISKEVLMGKDEYFQSF 860
AWLHKA+R+R YRE+I +NEV+L A D INEFGWDNKHAGINVLISKEVLMGK EYF+SF
Sbjct: 244 AWLHKASRKRAYREFIVKNEVILKAGDTINEFGWDNKHAGINVLISKEVLMGKAEYFESF 303
Query: 859 RVNADNFMCTLLPGISNHPQIQYSPGGLLFKVGSSNMQHVTQLSFLLLAYSNYLSHAGGR 680
+ NAD F+C++LPGIS HPQ+QYS GGLL K G SNMQHVT LSFLLLAYSNYLSHA
Sbjct: 304 KQNADGFICSILPGIS-HPQVQYSRGGLLVKTGGSNMQHVTSLSFLLLAYSNYLSHAKKV 362
Query: 679 VSCGSSSASPVQLRRVAKRQVDYILGDNPLRMSYMVGYGARFPRRIHHRASSLPSVAAHP 500
V CG +ASP LR++AKRQVDYILGDNP+ +SYMVGYG +FPRRIHHR SS+PSV+AHP
Sbjct: 363 VPCGELTASPSLLRQIAKRQVDYILGDNPMGLSYMVGYGQKFPRRIHHRGSSVPSVSAHP 422
Query: 499 ARIGCKAGAAYYTSPAPNPNLLVGAVVGGPTDDSDAFPDARAVFQQSEPTTYINAPLMGL 320
+ IGCK G+ Y+ SP PNPNLLVGAVVGGP + +DAFPD+R FQQSEPTTYINAPL+GL
Sbjct: 423 SHIGCKEGSRYFLSPNPNPNLLVGAVVGGP-NVTDAFPDSRPYFQQSEPTTYINAPLVGL 481
Query: 319 LAYFSAH 299
L YFSAH
Sbjct: 482 LGYFSAH 488
Score = 84.7 bits (208), Expect(2) = e-124
Identities = 39/56 (69%), Positives = 46/56 (82%)
Frame = -2
Query: 1218 SIVFRDADPAYSKRLLDRAVAVFEFADKYRGPYSSSLHAAVCPCYCDYSGYQDELL 1051
SIVFR DPAYS+ LLDRA VF FA++YRG YS+SL+ A CP YCD++GYQDELL
Sbjct: 185 SIVFRKRDPAYSRLLLDRATRVFAFANRYRGAYSNSLYHADCPFYCDFNGYQDELL 240
>sptr|Q96547|Q96547 Cellulase (EC 3.2.1.4).
Length = 485
Score = 375 bits (964), Expect(2) = e-123
Identities = 182/247 (73%), Positives = 209/247 (84%)
Frame = -1
Query: 1039 AWLHKATRRREYREYIKRNEVVLGASDAINEFGWDNKHAGINVLISKEVLMGKDEYFQSF 860
AWLHKATRRR+YREYI +NEV+L A D INEFGWDNKHAGINVLISKEVLMG+ +SF
Sbjct: 240 AWLHKATRRRQYREYIVKNEVILRAGDTINEFGWDNKHAGINVLISKEVLMGRAPDLKSF 299
Query: 859 RVNADNFMCTLLPGISNHPQIQYSPGGLLFKVGSSNMQHVTQLSFLLLAYSNYLSHAGGR 680
+VNAD F+C++LPGI+ HPQ+QYSPGGL+ K G NMQHVT LSFL LAYSNYLSHA
Sbjct: 300 QVNADAFICSILPGIA-HPQVQYSPGGLIVKPGVCNMQHVTSLSFLFLAYSNYLSHANHV 358
Query: 679 VSCGSSSASPVQLRRVAKRQVDYILGDNPLRMSYMVGYGARFPRRIHHRASSLPSVAAHP 500
V CGS SA+P L+ +AKRQVDYILGDNP RMSYMVGYG +P RIHHR SSLPS+AAH
Sbjct: 359 VPCGSMSATPALLKHIAKRQVDYILGDNPQRMSYMVGYGPHYPLRIHHRGSSLPSMAAHS 418
Query: 499 ARIGCKAGAAYYTSPAPNPNLLVGAVVGGPTDDSDAFPDARAVFQQSEPTTYINAPLMGL 320
ARIGCK G+ Y+ SP PNPN L+GAVVGGP + +D+FPDAR+ FQ+SEPTTY+NAPL+GL
Sbjct: 419 ARIGCKEGSRYFFSPNPNPNRLIGAVVGGP-NLTDSFPDARSFFQESEPTTYVNAPLVGL 477
Query: 319 LAYFSAH 299
LAYFSAH
Sbjct: 478 LAYFSAH 484
Score = 89.4 bits (220), Expect(2) = e-123
Identities = 41/56 (73%), Positives = 48/56 (85%)
Frame = -2
Query: 1218 SIVFRDADPAYSKRLLDRAVAVFEFADKYRGPYSSSLHAAVCPCYCDYSGYQDELL 1051
SIVFR D +YS LLDRAV VFEFA+++RG YSSSLH+AVCP YCD++GYQDELL
Sbjct: 181 SIVFRSLDASYSNLLLDRAVKVFEFANRHRGAYSSSLHSAVCPFYCDFNGYQDELL 236
>sptr|Q42872|Q42872 Endo-1,4-beta-glucanase precursor (EC 3.2.1.4).
Length = 489
Score = 376 bits (966), Expect(2) = e-123
Identities = 182/249 (73%), Positives = 209/249 (83%)
Frame = -1
Query: 1039 AWLHKATRRREYREYIKRNEVVLGASDAINEFGWDNKHAGINVLISKEVLMGKDEYFQSF 860
AWLHKATRRR+YREYI +NEV+L A D INEFGWDNKHAGINVLISKEVLMGK +SF
Sbjct: 243 AWLHKATRRRQYREYIVKNEVILRAGDTINEFGWDNKHAGINVLISKEVLMGKAPDLKSF 302
Query: 859 RVNADNFMCTLLPGISNHPQIQYSPGGLLFKVGSSNMQHVTQLSFLLLAYSNYLSHAGGR 680
+VNAD F+C++LPGIS HPQ+QYSPGGL+ K G NMQHVT LSFLLL YSNYLSHA
Sbjct: 303 QVNADAFICSILPGIS-HPQVQYSPGGLIVKPGVCNMQHVTSLSFLLLTYSNYLSHANHV 361
Query: 679 VSCGSSSASPVQLRRVAKRQVDYILGDNPLRMSYMVGYGARFPRRIHHRASSLPSVAAHP 500
V CGS +A+P L+ +AKRQVDYILGDNP RMSYMVGYG +P+RIHHR SS+PSVA H
Sbjct: 362 VPCGSMTATPALLKHIAKRQVDYILGDNPQRMSYMVGYGPHYPQRIHHRGSSVPSVATHS 421
Query: 499 ARIGCKAGAAYYTSPAPNPNLLVGAVVGGPTDDSDAFPDARAVFQQSEPTTYINAPLMGL 320
ARIGCK G+ Y+ SP PNPN L+GAVVGGP + +D+FPDAR FQ+SEPTTY+NAPL+GL
Sbjct: 422 ARIGCKEGSRYFFSPNPNPNRLIGAVVGGP-NLTDSFPDARPYFQESEPTTYVNAPLVGL 480
Query: 319 LAYFSAHPN 293
LAYF+AH N
Sbjct: 481 LAYFAAHSN 489
Score = 88.2 bits (217), Expect(2) = e-123
Identities = 40/56 (71%), Positives = 48/56 (85%)
Frame = -2
Query: 1218 SIVFRDADPAYSKRLLDRAVAVFEFADKYRGPYSSSLHAAVCPCYCDYSGYQDELL 1051
SIVFR D +YS LLDRAV VF+FA+++RG YSSSLH+AVCP YCD++GYQDELL
Sbjct: 184 SIVFRSLDSSYSNLLLDRAVKVFDFANRHRGAYSSSLHSAVCPFYCDFNGYQDELL 239
>sptr|O49296|O49296 T26J12.2 protein (F26F24.6).
Length = 489
Score = 368 bits (945), Expect(2) = e-119
Identities = 178/247 (72%), Positives = 207/247 (83%)
Frame = -1
Query: 1039 AWLHKATRRREYREYIKRNEVVLGASDAINEFGWDNKHAGINVLISKEVLMGKDEYFQSF 860
AWLHKA+++R YRE+I +N+V+L A D I+EFGWDNKHAGINVL+SK VLMGK EYFQSF
Sbjct: 244 AWLHKASKKRVYREFIVKNQVILRAGDTIHEFGWDNKHAGINVLVSKMVLMGKAEYFQSF 303
Query: 859 RVNADNFMCTLLPGISNHPQIQYSPGGLLFKVGSSNMQHVTQLSFLLLAYSNYLSHAGGR 680
+ NAD F+C+LLPGIS HPQ+QYS GGLL K G SNMQHVT LSFLLL YSNYLSHA
Sbjct: 304 KQNADEFICSLLPGIS-HPQVQYSQGGLLVKSGGSNMQHVTSLSFLLLTYSNYLSHANKV 362
Query: 679 VSCGSSSASPVQLRRVAKRQVDYILGDNPLRMSYMVGYGARFPRRIHHRASSLPSVAAHP 500
V CG +ASP LR+VAKRQVDYILGDNP++MSYMVGYG+RFP++IHHR SS+PSV HP
Sbjct: 363 VPCGEFTASPALLRQVAKRQVDYILGDNPMKMSYMVGYGSRFPQKIHHRGSSVPSVVDHP 422
Query: 499 ARIGCKAGAAYYTSPAPNPNLLVGAVVGGPTDDSDAFPDARAVFQQSEPTTYINAPLMGL 320
RIGCK G+ Y+ S PNPNLL+GAVVGGP + +D FPD+R FQ +EPTTYINAPL+GL
Sbjct: 423 DRIGCKDGSRYFFSNNPNPNLLIGAVVGGP-NITDDFPDSRPYFQLTEPTTYINAPLLGL 481
Query: 319 LAYFSAH 299
L YFSAH
Sbjct: 482 LGYFSAH 488
Score = 85.1 bits (209), Expect(2) = e-119
Identities = 39/56 (69%), Positives = 43/56 (76%)
Frame = -2
Query: 1218 SIVFRDADPAYSKRLLDRAVAVFEFADKYRGPYSSSLHAAVCPCYCDYSGYQDELL 1051
SIVF DP YSK LLDRA VF FA KYRG YS SL+ AVCP YCD++GY+DELL
Sbjct: 185 SIVFEKRDPVYSKMLLDRATRVFAFAQKYRGAYSDSLYQAVCPFYCDFNGYEDELL 240
>sptr|O23696|O23696 Endo-1,4-beta-glucanase.
Length = 493
Score = 382 bits (981), Expect(2) = e-108
Identities = 185/247 (74%), Positives = 211/247 (85%)
Frame = -1
Query: 1039 AWLHKATRRREYREYIKRNEVVLGASDAINEFGWDNKHAGINVLISKEVLMGKDEYFQSF 860
AWLHKA+R+R YRE+I +NEV+L A D INEFGWDNKHAGINVLISKEVLMGK EYF+SF
Sbjct: 245 AWLHKASRKRAYREFIVKNEVILKAGDTINEFGWDNKHAGINVLISKEVLMGKAEYFESF 304
Query: 859 RVNADNFMCTLLPGISNHPQIQYSPGGLLFKVGSSNMQHVTQLSFLLLAYSNYLSHAGGR 680
+ NAD F+C++LPGIS HPQ+QYS GGLL K G SNMQHVT LSFLLLAYSNYLSHA
Sbjct: 305 KQNADGFICSILPGIS-HPQVQYSRGGLLVKTGGSNMQHVTSLSFLLLAYSNYLSHAKKV 363
Query: 679 VSCGSSSASPVQLRRVAKRQVDYILGDNPLRMSYMVGYGARFPRRIHHRASSLPSVAAHP 500
V CG +ASP LR++AKRQVDYILGDNP+ +SYMVGYG +FPRRIHHR SS+PSV+AHP
Sbjct: 364 VPCGELTASPSLLRQIAKRQVDYILGDNPMGLSYMVGYGQKFPRRIHHRGSSVPSVSAHP 423
Query: 499 ARIGCKAGAAYYTSPAPNPNLLVGAVVGGPTDDSDAFPDARAVFQQSEPTTYINAPLMGL 320
+ IGCK G+ Y+ SP PNPNL VGAVVGGP + +DAFPD+R FQQSEPTTYINAPL+GL
Sbjct: 424 SHIGCKEGSRYFLSPNPNPNLWVGAVVGGP-NVTDAFPDSRPYFQQSEPTTYINAPLVGL 482
Query: 319 LAYFSAH 299
L YFSAH
Sbjct: 483 LGYFSAH 489
Score = 33.5 bits (75), Expect(2) = e-108
Identities = 16/22 (72%), Positives = 17/22 (77%)
Frame = -2
Query: 1218 SIVFRDADPAYSKRLLDRAVAV 1153
SIVFR DPAYS+ LLDRA V
Sbjct: 185 SIVFRKRDPAYSRLLLDRATRV 206
>sptr|Q9SRX3|Q9SRX3 Endo-1,4-beta glucanase.
Length = 501
Score = 288 bits (738), Expect(2) = 2e-96
Identities = 141/245 (57%), Positives = 178/245 (72%)
Frame = -1
Query: 1039 AWLHKATRRREYREYIKRNEVVLGASDAINEFGWDNKHAGINVLISKEVLMGKDEYFQSF 860
AWL KAT Y YIK N +LGA + N F WDNKH G +L+SKE L+ K + + +
Sbjct: 259 AWLQKATNNPTYLNYIKANGQILGADEFDNMFSWDNKHVGARILLSKEFLIQKVKSLEEY 318
Query: 859 RVNADNFMCTLLPGISNHPQIQYSPGGLLFKVGSSNMQHVTQLSFLLLAYSNYLSHAGGR 680
+ +AD+F+C++LPG S+ QY+PGGLLFK+G SNMQ+VT SFLLL Y+ YL+ A
Sbjct: 319 KEHADSFICSVLPGASSS---QYTPGGLLFKMGESNMQYVTSTSFLLLTYAKYLTSARTV 375
Query: 679 VSCGSSSASPVQLRRVAKRQVDYILGDNPLRMSYMVGYGARFPRRIHHRASSLPSVAAHP 500
CG S +P +LR +AK+QVDY+LG NPL+MSYMVGYG ++PRRIHHR SSLPSVA HP
Sbjct: 376 AYCGGSVVTPARLRSIAKKQVDYLLGGNPLKMSYMVGYGLKYPRRIHHRGSSLPSVAVHP 435
Query: 499 ARIGCKAGAAYYTSPAPNPNLLVGAVVGGPTDDSDAFPDARAVFQQSEPTTYINAPLMGL 320
RI C G + +TS +PNPN LVGAVVGGP D +D FPD R+ + +SEP TYINAPL+G
Sbjct: 436 TRIQCHDGFSLFTSQSPNPNDLVGAVVGGP-DQNDQFPDERSDYGRSEPATYINAPLVGA 494
Query: 319 LAYFS 305
LAY +
Sbjct: 495 LAYLA 499
Score = 87.8 bits (216), Expect(2) = 2e-96
Identities = 40/56 (71%), Positives = 43/56 (76%)
Frame = -2
Query: 1218 SIVFRDADPAYSKRLLDRAVAVFEFADKYRGPYSSSLHAAVCPCYCDYSGYQDELL 1051
SIVFR DP+YS LL RA+ VF FADKYRGPYS+ L VCP YC YSGYQDELL
Sbjct: 200 SIVFRKCDPSYSNHLLQRAITVFTFADKYRGPYSAGLAPEVCPFYCSYSGYQDELL 255
>sptr|O81416|O81416 T2H3.5 protein (Putative endo-1, 4-beta glucanase)
(AT4g02290/T2H3_5).
Length = 516
Score = 291 bits (744), Expect(2) = 3e-96
Identities = 142/245 (57%), Positives = 180/245 (73%)
Frame = -1
Query: 1039 AWLHKATRRREYREYIKRNEVVLGASDAINEFGWDNKHAGINVLISKEVLMGKDEYFQSF 860
AWL KAT+ +Y YIK N +LGA++ N FGWDNKHAG +L++K L+ + +
Sbjct: 268 AWLQKATKNIKYLNYIKINGQILGAAEYDNTFGWDNKHAGARILLTKAFLVQNVKTLHEY 327
Query: 859 RVNADNFMCTLLPGISNHPQIQYSPGGLLFKVGSSNMQHVTQLSFLLLAYSNYLSHAGGR 680
+ +ADNF+C+++PG + QY+PGGLLFK+ +NMQ+VT SFLLL Y+ YL+ A
Sbjct: 328 KGHADNFICSVIPG-APFSSTQYTPGGLLFKMADANMQYVTSTSFLLLTYAKYLTSAKTV 386
Query: 679 VSCGSSSASPVQLRRVAKRQVDYILGDNPLRMSYMVGYGARFPRRIHHRASSLPSVAAHP 500
V CG S +P +LR +AKRQVDY+LGDNPLRMSYMVGYG +FPRRIHHR SSLP VA+HP
Sbjct: 387 VHCGGSVYTPGRLRSIAKRQVDYLLGDNPLRMSYMVGYGPKFPRRIHHRGSSLPCVASHP 446
Query: 499 ARIGCKAGAAYYTSPAPNPNLLVGAVVGGPTDDSDAFPDARAVFQQSEPTTYINAPLMGL 320
A+I C G A S +PNPN LVGAVVGGP D D FPD R+ ++QSEP TYIN+PL+G
Sbjct: 447 AKIQCHQGFAIMNSQSPNPNFLVGAVVGGP-DQHDRFPDERSDYEQSEPATYINSPLVGA 505
Query: 319 LAYFS 305
LAYF+
Sbjct: 506 LAYFA 510
Score = 84.7 bits (208), Expect(2) = 3e-96
Identities = 39/56 (69%), Positives = 45/56 (80%)
Frame = -2
Query: 1218 SIVFRDADPAYSKRLLDRAVAVFEFADKYRGPYSSSLHAAVCPCYCDYSGYQDELL 1051
+IVFR +DP+YSK LL RA++VF FADKYRG YS+ L VCP YC YSGYQDELL
Sbjct: 209 AIVFRKSDPSYSKVLLKRAISVFAFADKYRGTYSAGLKPDVCPFYCSYSGYQDELL 264
>sptr|O64949|O64949 Endo-1,4-beta glucanase.
Length = 501
Score = 288 bits (736), Expect(2) = 3e-96
Identities = 140/245 (57%), Positives = 178/245 (72%)
Frame = -1
Query: 1039 AWLHKATRRREYREYIKRNEVVLGASDAINEFGWDNKHAGINVLISKEVLMGKDEYFQSF 860
AWL KAT Y YIK N +LGA + N F WDNKH G +L+SKE L+ K + + +
Sbjct: 259 AWLQKATNNPTYLNYIKANGQILGADEFDNMFSWDNKHVGARILLSKEFLIQKVKSLEEY 318
Query: 859 RVNADNFMCTLLPGISNHPQIQYSPGGLLFKVGSSNMQHVTQLSFLLLAYSNYLSHAGGR 680
+ +AD+F+C+++PG S+ QY+PGGLLFK+G SNMQ+VT SFLLL Y+ YL+ A
Sbjct: 319 KEHADSFICSVIPGASSS---QYTPGGLLFKMGESNMQYVTSTSFLLLTYAKYLTSARTV 375
Query: 679 VSCGSSSASPVQLRRVAKRQVDYILGDNPLRMSYMVGYGARFPRRIHHRASSLPSVAAHP 500
CG S +P +LR +AK+QVDY+LG NPL+MSYMVGYG ++PRRIHHR SSLPSVA HP
Sbjct: 376 AYCGGSVVTPARLRSIAKKQVDYLLGGNPLKMSYMVGYGLKYPRRIHHRGSSLPSVAVHP 435
Query: 499 ARIGCKAGAAYYTSPAPNPNLLVGAVVGGPTDDSDAFPDARAVFQQSEPTTYINAPLMGL 320
RI C G + +TS +PNPN LVGAVVGGP D +D FPD R+ + +SEP TYINAPL+G
Sbjct: 436 TRIQCHDGFSLFTSQSPNPNDLVGAVVGGP-DQNDQFPDERSDYGRSEPATYINAPLVGA 494
Query: 319 LAYFS 305
LAY +
Sbjct: 495 LAYLA 499
Score = 87.8 bits (216), Expect(2) = 3e-96
Identities = 40/56 (71%), Positives = 43/56 (76%)
Frame = -2
Query: 1218 SIVFRDADPAYSKRLLDRAVAVFEFADKYRGPYSSSLHAAVCPCYCDYSGYQDELL 1051
SIVFR DP+YS LL RA+ VF FADKYRGPYS+ L VCP YC YSGYQDELL
Sbjct: 200 SIVFRKCDPSYSNHLLQRAITVFTFADKYRGPYSAGLAPEVCPFYCSYSGYQDELL 255
>sptr|Q42875|Q42875 Endo-1,4-beta-glucanase precursor (EC 3.2.1.4).
Length = 510
Score = 293 bits (750), Expect(2) = 6e-95
Identities = 141/245 (57%), Positives = 182/245 (74%)
Frame = -1
Query: 1039 AWLHKATRRREYREYIKRNEVVLGASDAINEFGWDNKHAGINVLISKEVLMGKDEYFQSF 860
AWLH+AT+ Y YI+RN LGA++ N FGWDNKH G +L+SK L+ K + +
Sbjct: 262 AWLHRATKNPTYLNYIQRNGQTLGAAETDNTFGWDNKHVGARILLSKSFLVQKLQTLHDY 321
Query: 859 RVNADNFMCTLLPGISNHPQIQYSPGGLLFKVGSSNMQHVTQLSFLLLAYSNYLSHAGGR 680
+ +ADN++C+L+PG + Q QY+PGGLLFK+ SNMQ+VT SFLL+ Y+ YL+ A
Sbjct: 322 KSHADNYICSLIPG-TPASQAQYTPGGLLFKMDDSNMQYVTSTSFLLVTYAKYLTSARMV 380
Query: 679 VSCGSSSASPVQLRRVAKRQVDYILGDNPLRMSYMVGYGARFPRRIHHRASSLPSVAAHP 500
V CG +P +LR VAK+QVDY+LGDNPL+MSYMVGYGAR+P+RIHHR SSLPSVA HP
Sbjct: 381 VKCGGVVITPKRLRNVAKKQVDYLLGDNPLKMSYMVGYGARYPQRIHHRGSSLPSVANHP 440
Query: 499 ARIGCKAGAAYYTSPAPNPNLLVGAVVGGPTDDSDAFPDARAVFQQSEPTTYINAPLMGL 320
A+I C+ G + S +PNPN+LVGAVVGGP D+ D FPD R+ ++QSEP TYINAPL+G
Sbjct: 441 AKIQCRDGFSVMNSQSPNPNVLVGAVVGGP-DEHDRFPDERSDYEQSEPATYINAPLVGT 499
Query: 319 LAYFS 305
L Y +
Sbjct: 500 LTYLA 504
Score = 78.2 bits (191), Expect(2) = 6e-95
Identities = 35/56 (62%), Positives = 42/56 (75%)
Frame = -2
Query: 1218 SIVFRDADPAYSKRLLDRAVAVFEFADKYRGPYSSSLHAAVCPCYCDYSGYQDELL 1051
S+VFR +P+YSK L+ RA+ VF FADKYRG YS+ L VCP YC SGY+DELL
Sbjct: 203 SLVFRKCNPSYSKILIKRAIRVFAFADKYRGSYSNGLRKVVCPYYCSVSGYEDELL 258
>sptr|O64401|O64401 Endo-beta-1,4-glucanase.
Length = 510
Score = 285 bits (730), Expect(2) = 2e-94
Identities = 142/245 (57%), Positives = 176/245 (71%)
Frame = -1
Query: 1039 AWLHKATRRREYREYIKRNEVVLGASDAINEFGWDNKHAGINVLISKEVLMGKDEYFQSF 860
AWLHKATR Y YI+ LGA ++ N FGWDNKHAG VL+SKE L+ + +
Sbjct: 263 AWLHKATRNPTYLNYIQSKGQSLGADESDNIFGWDNKHAGARVLLSKEFLLRNVKSLHDY 322
Query: 859 RVNADNFMCTLLPGISNHPQIQYSPGGLLFKVGSSNMQHVTQLSFLLLAYSNYLSHAGGR 680
+ +ADN++C+LLPG+S + Q +Y+PGGLL+ + SN+Q+VT SFLL Y+ YLS +
Sbjct: 323 KGHADNYICSLLPGVS-YSQAKYTPGGLLYTLSDSNLQYVTSSSFLLFTYAKYLSSSKHV 381
Query: 679 VSCGSSSASPVQLRRVAKRQVDYILGDNPLRMSYMVGYGARFPRRIHHRASSLPSVAAHP 500
V+CGS + SP +LR +AKRQVDYILGDNPL+MSYMVGYG+ +P RIHHR SSLPS HP
Sbjct: 382 VTCGSMTFSPKRLRTIAKRQVDYILGDNPLKMSYMVGYGSHYPERIHHRGSSLPSKGNHP 441
Query: 499 ARIGCKAGAAYYTSPAPNPNLLVGAVVGGPTDDSDAFPDARAVFQQSEPTTYINAPLMGL 320
I C AG S APNPN+LVGAVVGGP D D FPD R ++QSEPTTYINAP +G
Sbjct: 442 QAIPCNAGFQSLYSNAPNPNILVGAVVGGP-DSMDRFPDDRNDYEQSEPTTYINAPFVGS 500
Query: 319 LAYFS 305
LAY +
Sbjct: 501 LAYLA 505
Score = 84.0 bits (206), Expect(2) = 2e-94
Identities = 37/56 (66%), Positives = 45/56 (80%)
Frame = -2
Query: 1218 SIVFRDADPAYSKRLLDRAVAVFEFADKYRGPYSSSLHAAVCPCYCDYSGYQDELL 1051
S+VFR +DP YSKRL+ A+ VF+FADK+RG YS SL + VCP YC YSGY+DELL
Sbjct: 204 SLVFRKSDPQYSKRLIHTAMRVFDFADKHRGAYSDSLRSCVCPFYCSYSGYEDELL 259
>sptr|Q9SSU7|Q9SSU7 Endo-1,4-beta-glucanase.
Length = 506
Score = 290 bits (742), Expect(2) = 4e-93
Identities = 135/245 (55%), Positives = 182/245 (74%)
Frame = -1
Query: 1039 AWLHKATRRREYREYIKRNEVVLGASDAINEFGWDNKHAGINVLISKEVLMGKDEYFQSF 860
AWLHKAT+ Y +YI+ N +LGA++ N FGWDNKH G +L+SKE L+ + +
Sbjct: 258 AWLHKATKNPMYLKYIQTNGQILGAAEFDNTFGWDNKHVGARILLSKEFLVQNVKSLHDY 317
Query: 859 RVNADNFMCTLLPGISNHPQIQYSPGGLLFKVGSSNMQHVTQLSFLLLAYSNYLSHAGGR 680
+ ++DNF+C+L+PG + QY+PGGLLFK+ SNMQ+VT +FLL+ Y+ YL+ +
Sbjct: 318 KGHSDNFVCSLIPGAGSS-SAQYTPGGLLFKMSDSNMQYVTSTTFLLVTYAKYLTKSHSV 376
Query: 679 VSCGSSSASPVQLRRVAKRQVDYILGDNPLRMSYMVGYGARFPRRIHHRASSLPSVAAHP 500
V+CG ++ +P +LR +AKRQVDY+LGDNPL+MSYMVGYG R+P+RIHHR SSLPS+A HP
Sbjct: 377 VNCGGTTVTPKRLRTLAKRQVDYLLGDNPLKMSYMVGYGPRYPQRIHHRGSSLPSMAVHP 436
Query: 499 ARIGCKAGAAYYTSPAPNPNLLVGAVVGGPTDDSDAFPDARAVFQQSEPTTYINAPLMGL 320
+I C AG S +PNPN+L+GAVVGGP D D FPD R+ ++QSEP TY+NAPL+G
Sbjct: 437 GKIQCSAGFGVMNSKSPNPNILMGAVVGGP-DQHDRFPDQRSDYEQSEPATYVNAPLVGT 495
Query: 319 LAYFS 305
LAY +
Sbjct: 496 LAYLA 500
Score = 75.1 bits (183), Expect(2) = 4e-93
Identities = 34/56 (60%), Positives = 43/56 (76%)
Frame = -2
Query: 1218 SIVFRDADPAYSKRLLDRAVAVFEFADKYRGPYSSSLHAAVCPCYCDYSGYQDELL 1051
S+VFR +DP Y+K L+ RA+ VF+FADK+R YS++L VCP YC YSGYQD LL
Sbjct: 199 SLVFRKSDPTYAKILVRRAIRVFQFADKHRRSYSNALKPFVCPFYCSYSGYQDGLL 254
>sptr|Q8LQ92|Q8LQ92 Putative endo-1,3(4)-beta-glucanase.
Length = 499
Score = 282 bits (722), Expect(2) = 8e-93
Identities = 139/245 (56%), Positives = 177/245 (72%)
Frame = -1
Query: 1039 AWLHKATRRREYREYIKRNEVVLGASDAINEFGWDNKHAGINVLISKEVLMGKDEYFQSF 860
AWLH+AT+ Y YI+ N VLGA + N FGWDNKHAG +LI+K L+ K +
Sbjct: 251 AWLHRATKNPTYLSYIQMNGQVLGADEQDNTFGWDNKHAGARILIAKAFLVQKVAALHEY 310
Query: 859 RVNADNFMCTLLPGISNHPQIQYSPGGLLFKVGSSNMQHVTQLSFLLLAYSNYLSHAGGR 680
+ +AD+F+C+++PG Q QY+ GGLLFK+ SNMQ+VT SFLLL Y+ YL+ +
Sbjct: 311 KGHADSFICSMVPGTPTD-QTQYTRGGLLFKLSDSNMQYVTSSSFLLLTYAKYLAFSKTT 369
Query: 679 VSCGSSSASPVQLRRVAKRQVDYILGDNPLRMSYMVGYGARFPRRIHHRASSLPSVAAHP 500
VSCG ++ +P +LR +A++QVDY+LG NP+ MSYMVGYGA++PRRIHHRASSLPSVAAHP
Sbjct: 370 VSCGGAAVTPARLRAIARQQVDYLLGSNPMGMSYMVGYGAKYPRRIHHRASSLPSVAAHP 429
Query: 499 ARIGCKAGAAYYTSPAPNPNLLVGAVVGGPTDDSDAFPDARAVFQQSEPTTYINAPLMGL 320
ARIGC G S NPN+LVGAVVGGP + D FPD R+ + SEP TYINAPL+G
Sbjct: 430 ARIGCSQGFTALYSGVANPNVLVGAVVGGP-NLQDQFPDQRSDHEHSEPATYINAPLVGA 488
Query: 319 LAYFS 305
LAY +
Sbjct: 489 LAYLA 493
Score = 82.0 bits (201), Expect(2) = 8e-93
Identities = 38/56 (67%), Positives = 44/56 (78%)
Frame = -2
Query: 1218 SIVFRDADPAYSKRLLDRAVAVFEFADKYRGPYSSSLHAAVCPCYCDYSGYQDELL 1051
S+VFR +DPAY+ RL+ RA VFEFADK+RG YS+ L VCP YC YSGYQDELL
Sbjct: 192 SLVFRKSDPAYASRLVARAKRVFEFADKHRGTYSTRLSPYVCPYYCSYSGYQDELL 247
>sptr|Q9AYM9|Q9AYM9 Beta-1,4-glucanase.
Length = 493
Score = 273 bits (697), Expect(2) = 5e-92
Identities = 133/247 (53%), Positives = 175/247 (70%)
Frame = -1
Query: 1039 AWLHKATRRREYREYIKRNEVVLGASDAINEFGWDNKHAGINVLISKEVLMGKDEYFQSF 860
AW+HKA+ Y YI+ N +LGA D F WD+K AG VL++K+ L + E FQ +
Sbjct: 245 AWIHKASGDSSYLSYIQSNGHILGAEDDDFSFSWDDKKAGTKVLLAKDFLQDRVEQFQVY 304
Query: 859 RVNADNFMCTLLPGISNHPQIQYSPGGLLFKVGSSNMQHVTQLSFLLLAYSNYLSHAGGR 680
+ +ADNF+C+ +PG +++ Q QY+PGGLLFK SN+Q+VT SFLL+AY+ YL G
Sbjct: 305 KAHADNFICSFIPGANDY-QAQYTPGGLLFKQSDSNLQYVTTTSFLLVAYAKYLGKNGDI 363
Query: 679 VSCGSSSASPVQLRRVAKRQVDYILGDNPLRMSYMVGYGARFPRRIHHRASSLPSVAAHP 500
+CGS+ + +L VA+ QVDYILGDNP MSYMVG+G ++P+ IHHRASSLPSV AHP
Sbjct: 364 TTCGSTVITAKKLISVAREQVDYILGDNPATMSYMVGFGNKYPQHIHHRASSLPSVHAHP 423
Query: 499 ARIGCKAGAAYYTSPAPNPNLLVGAVVGGPTDDSDAFPDARAVFQQSEPTTYINAPLMGL 320
+RIGC G Y S +PNPN+LVGA++GGP D D F D R ++QSEP TYINAP +G
Sbjct: 424 SRIGCNDGFQYLNSGSPNPNVLVGAILGGP-DSGDKFTDDRNNYRQSEPATYINAPFVGA 482
Query: 319 LAYFSAH 299
A+FSA+
Sbjct: 483 AAFFSAY 489
Score = 89.0 bits (219), Expect(2) = 5e-92
Identities = 39/56 (69%), Positives = 47/56 (83%)
Frame = -2
Query: 1218 SIVFRDADPAYSKRLLDRAVAVFEFADKYRGPYSSSLHAAVCPCYCDYSGYQDELL 1051
SIVF+D+DP+YS +LL A+ VF+FADKYRG YS SL++ VCP YC YSGYQDELL
Sbjct: 186 SIVFKDSDPSYSSKLLHTAIQVFDFADKYRGSYSDSLNSVVCPFYCSYSGYQDELL 241
>sptr|Q93YQ7|Q93YQ7 Putative endo-1, 4-beta-glucanase.
Length = 497
Score = 287 bits (734), Expect(2) = 2e-91
Identities = 134/248 (54%), Positives = 184/248 (74%)
Frame = -1
Query: 1039 AWLHKATRRREYREYIKRNEVVLGASDAINEFGWDNKHAGINVLISKEVLMGKDEYFQSF 860
AWL KAT Y YI+ N GA+D ++EFGWDNK G+NVL+SKEV+ G +++
Sbjct: 249 AWLRKATGDESYLNYIESNREPFGANDNVDEFGWDNKVGGLNVLVSKEVIEGNMYNLEAY 308
Query: 859 RVNADNFMCTLLPGISNHPQIQYSPGGLLFKVGSSNMQHVTQLSFLLLAYSNYLSHAGGR 680
+ +A++FMC+L+P S+ P ++Y+ GLL+K G S +QH T +SFLLL Y+ YLS +
Sbjct: 309 KASAESFMCSLVPE-SSGPHVEYTSAGLLYKPGGSQLQHATTISFLLLVYAQYLSRSSLS 367
Query: 679 VSCGSSSASPVQLRRVAKRQVDYILGDNPLRMSYMVGYGARFPRRIHHRASSLPSVAAHP 500
++CG+ + P LRR+AK+QVDYILG+NP+ +SYMVGYG R+P+RIHHR SSLPS+ HP
Sbjct: 368 LNCGTLTVPPDYLRRLAKKQVDYILGNNPMGLSYMVGYGERYPKRIHHRGSSLPSIVDHP 427
Query: 499 ARIGCKAGAAYYTSPAPNPNLLVGAVVGGPTDDSDAFPDARAVFQQSEPTTYINAPLMGL 320
I CK G+ Y+ S PNPN+L+GAVVGGP +D D + D R+ F++SEPTTYINAP +G+
Sbjct: 428 EAIRCKDGSVYFNSTEPNPNVLIGAVVGGPGED-DMYDDDRSDFRKSEPTTYINAPFVGV 486
Query: 319 LAYFSAHP 296
LAYF+A+P
Sbjct: 487 LAYFAANP 494
Score = 72.4 bits (176), Expect(2) = 2e-91
Identities = 34/58 (58%), Positives = 42/58 (72%), Gaps = 2/58 (3%)
Frame = -2
Query: 1218 SIVFRDADPAYSKRLLDRAVAVFEFADKYRGPYSSS--LHAAVCPCYCDYSGYQDELL 1051
SI FR +DP YS+ LL AV F+FAD YRG YSS+ + VCP YCD++G+QDELL
Sbjct: 188 SIAFRSSDPGYSQTLLQNAVKTFQFADMYRGAYSSNDDIKNDVCPFYCDFNGFQDELL 245
>sptr|Q9SVJ2|Q9SVJ2 Putative endo-1, 4-beta-glucanase.
Length = 475
Score = 287 bits (734), Expect(2) = 2e-91
Identities = 134/248 (54%), Positives = 184/248 (74%)
Frame = -1
Query: 1039 AWLHKATRRREYREYIKRNEVVLGASDAINEFGWDNKHAGINVLISKEVLMGKDEYFQSF 860
AWL KAT Y YI+ N GA+D ++EFGWDNK G+NVL+SKEV+ G +++
Sbjct: 227 AWLRKATGDESYLNYIESNREPFGANDNVDEFGWDNKVGGLNVLVSKEVIEGNMYNLEAY 286
Query: 859 RVNADNFMCTLLPGISNHPQIQYSPGGLLFKVGSSNMQHVTQLSFLLLAYSNYLSHAGGR 680
+ +A++FMC+L+P S+ P ++Y+ GLL+K G S +QH T +SFLLL Y+ YLS +
Sbjct: 287 KASAESFMCSLVPE-SSGPHVEYTSAGLLYKPGGSQLQHATTISFLLLVYAQYLSRSSLS 345
Query: 679 VSCGSSSASPVQLRRVAKRQVDYILGDNPLRMSYMVGYGARFPRRIHHRASSLPSVAAHP 500
++CG+ + P LRR+AK+QVDYILG+NP+ +SYMVGYG R+P+RIHHR SSLPS+ HP
Sbjct: 346 LNCGTLTVPPDYLRRLAKKQVDYILGNNPMGLSYMVGYGERYPKRIHHRGSSLPSIVDHP 405
Query: 499 ARIGCKAGAAYYTSPAPNPNLLVGAVVGGPTDDSDAFPDARAVFQQSEPTTYINAPLMGL 320
I CK G+ Y+ S PNPN+L+GAVVGGP +D D + D R+ F++SEPTTYINAP +G+
Sbjct: 406 EAIRCKDGSVYFNSTEPNPNVLIGAVVGGPGED-DMYDDDRSDFRKSEPTTYINAPFVGV 464
Query: 319 LAYFSAHP 296
LAYF+A+P
Sbjct: 465 LAYFAANP 472
Score = 72.4 bits (176), Expect(2) = 2e-91
Identities = 34/58 (58%), Positives = 42/58 (72%), Gaps = 2/58 (3%)
Frame = -2
Query: 1218 SIVFRDADPAYSKRLLDRAVAVFEFADKYRGPYSSS--LHAAVCPCYCDYSGYQDELL 1051
SI FR +DP YS+ LL AV F+FAD YRG YSS+ + VCP YCD++G+QDELL
Sbjct: 166 SIAFRSSDPGYSQTLLQNAVKTFQFADMYRGAYSSNDDIKNDVCPFYCDFNGFQDELL 223
>sw|P05522|GUN1_PERAE Endoglucanase 1 precursor (EC 3.2.1.4)
(Endo-1,4-beta-glucanase) (Abscission cellulase 1).
Length = 494
Score = 275 bits (703), Expect(2) = 3e-90
Identities = 131/248 (52%), Positives = 177/248 (71%)
Frame = -1
Query: 1039 AWLHKATRRREYREYIKRNEVVLGASDAINEFGWDNKHAGINVLISKEVLMGKDEYFQSF 860
+WLH+A++ Y YI+ N LGA D F WD+K G VL+SK L + E Q +
Sbjct: 246 SWLHRASQNASYMTYIQSNGHTLGADDDDYSFSWDDKRVGTKVLLSKGFLQDRIEELQLY 305
Query: 859 RVNADNFMCTLLPGISNHPQIQYSPGGLLFKVGSSNMQHVTQLSFLLLAYSNYLSHAGGR 680
+V+ DN++C+L+PG S+ Q QY+PGGLL+K +SN+Q+VT +FLLL Y+NYL+ +GG
Sbjct: 306 KVHTDNYICSLIPGTSSF-QAQYTPGGLLYKGSASNLQYVTSTAFLLLTYANYLNSSGGH 364
Query: 679 VSCGSSSASPVQLRRVAKRQVDYILGDNPLRMSYMVGYGARFPRRIHHRASSLPSVAAHP 500
SCG+++ + L +AK+QVDYILG NP +MSYMVG+G R+P+ +HHR SSLPSV HP
Sbjct: 365 ASCGTTTVTAKNLISLAKKQVDYILGQNPAKMSYMVGFGERYPQHVHHRGSSLPSVQVHP 424
Query: 499 ARIGCKAGAAYYTSPAPNPNLLVGAVVGGPTDDSDAFPDARAVFQQSEPTTYINAPLMGL 320
I C AG Y S PNPN+LVGA++GGP D+ D+F D R +QQSEP TYINAPL+G
Sbjct: 425 NSIPCNAGFQYLYSSPPNPNILVGAILGGP-DNRDSFSDDRNNYQQSEPATYINAPLVGA 483
Query: 319 LAYFSAHP 296
LA+F+A+P
Sbjct: 484 LAFFAANP 491
Score = 80.9 bits (198), Expect(2) = 3e-90
Identities = 38/56 (67%), Positives = 43/56 (76%)
Frame = -2
Query: 1218 SIVFRDADPAYSKRLLDRAVAVFEFADKYRGPYSSSLHAAVCPCYCDYSGYQDELL 1051
SIVF D+D +YS +LL AV VFEFAD+YRG YS SL + VCP YC YSGY DELL
Sbjct: 187 SIVFGDSDSSYSTKLLHTAVKVFEFADQYRGSYSDSLGSVVCPFYCSYSGYNDELL 242
>sptr|P94114|P94114 Endo-beta-1,4-glucanase (EC 3.2.1.4) (Cellulase)
(Endoglucanase) (Endo-1,4-beta-glucanase) (Carboxymethyl
cellulase).
Length = 497
Score = 273 bits (698), Expect(2) = 3e-90
Identities = 133/250 (53%), Positives = 173/250 (69%)
Frame = -1
Query: 1039 AWLHKATRRREYREYIKRNEVVLGASDAINEFGWDNKHAGINVLISKEVLMGKDEYFQSF 860
+W+H+A++ Y YIK N +LGA D F WD+K G VL+SK L +E FQ +
Sbjct: 249 SWIHRASQNSSYLAYIKSNGHILGADDDGFSFSWDDKRPGTKVLLSKNFLEKNNEEFQLY 308
Query: 859 RVNADNFMCTLLPGISNHPQIQYSPGGLLFKVGSSNMQHVTQLSFLLLAYSNYLSHAGGR 680
+ ++DN++C+LLPG SN Q QY+PGGLL+K SN+Q+VT + LLL Y+ YL GG
Sbjct: 309 KAHSDNYICSLLPGTSNF-QAQYTPGGLLYKASESNLQYVTSTTLLLLTYAKYLRTNGGV 367
Query: 679 VSCGSSSASPVQLRRVAKRQVDYILGDNPLRMSYMVGYGARFPRRIHHRASSLPSVAAHP 500
+CGSS + L AK+QVDYILG+NP ++SYMVG+G ++P IHHR SSLPSV HP
Sbjct: 368 ATCGSSKVTAETLISEAKKQVDYILGNNPAKISYMVGFGKKYPLHIHHRGSSLPSVHEHP 427
Query: 499 ARIGCKAGAAYYTSPAPNPNLLVGAVVGGPTDDSDAFPDARAVFQQSEPTTYINAPLMGL 320
RI C G Y S +PNPN+LVGA+VGGP D D+F D R +QQSEP TYINAP++G
Sbjct: 428 ERISCNNGFQYLNSGSPNPNVLVGAIVGGP-DSKDSFSDDRNNYQQSEPATYINAPIVGA 486
Query: 319 LAYFSAHPNP 290
LA+FSA+ NP
Sbjct: 487 LAFFSANTNP 496
Score = 82.4 bits (202), Expect(2) = 3e-90
Identities = 36/56 (64%), Positives = 45/56 (80%)
Frame = -2
Query: 1218 SIVFRDADPAYSKRLLDRAVAVFEFADKYRGPYSSSLHAAVCPCYCDYSGYQDELL 1051
SIVF+D+DP+YS +LL A+ VF+FAD+YRG YS S+ + VCP YC YSGY DELL
Sbjct: 190 SIVFKDSDPSYSGKLLHTAMKVFDFADRYRGSYSDSIGSVVCPFYCSYSGYHDELL 245
>sptr|O64402|O64402 Endo-beta-1,4-glucanase.
Length = 515
Score = 270 bits (691), Expect(2) = 1e-88
Identities = 136/245 (55%), Positives = 170/245 (69%)
Frame = -1
Query: 1039 AWLHKATRRREYREYIKRNEVVLGASDAINEFGWDNKHAGINVLISKEVLMGKDEYFQSF 860
AWL +A+R Y YI+ + LG D +N F WD+KHAG VL++KEVL+ + + +
Sbjct: 269 AWLQRASRNVSYISYIQSHGQNLGGEDNVNIFSWDDKHAGARVLLAKEVLLRNSKSLEEY 328
Query: 859 RVNADNFMCTLLPGISNHPQIQYSPGGLLFKVGSSNMQHVTQLSFLLLAYSNYLSHAGGR 680
R +ADNF+C+LLPG N Q QY+PGGLL+K+ N+Q+VT +FLL Y YL +
Sbjct: 329 RGHADNFVCSLLPGTPNS-QAQYTPGGLLYKMSDCNLQYVTSTTFLLFTYPKYLRVSKQV 387
Query: 679 VSCGSSSASPVQLRRVAKRQVDYILGDNPLRMSYMVGYGARFPRRIHHRASSLPSVAAHP 500
V CG+ +P ++R +AKRQVDYILGDNPLRMSYMVGYGA+FP RIHHR SSLPS+ HP
Sbjct: 388 VRCGNMVVTPTRIRTLAKRQVDYILGDNPLRMSYMVGYGAKFPERIHHRGSSLPSIYQHP 447
Query: 499 ARIGCKAGAAYYTSPAPNPNLLVGAVVGGPTDDSDAFPDARAVFQQSEPTTYINAPLMGL 320
I C G S APNPN LVGAVVGGP D++D F D R + SEPTTYINAPL+G
Sbjct: 448 QIIPCNDGFQSLYSNAPNPNRLVGAVVGGP-DNNDHFSDERNDYAHSEPTTYINAPLVGS 506
Query: 319 LAYFS 305
LAY +
Sbjct: 507 LAYLA 511
Score = 80.1 bits (196), Expect(2) = 1e-88
Identities = 36/56 (64%), Positives = 42/56 (75%)
Frame = -2
Query: 1218 SIVFRDADPAYSKRLLDRAVAVFEFADKYRGPYSSSLHAAVCPCYCDYSGYQDELL 1051
S+VFR DP YS+ LL A+ VF+FADK+RG YS SL + VCP YC YSGY DELL
Sbjct: 210 SMVFRTTDPIYSRHLLQTAMQVFDFADKHRGAYSDSLQSEVCPFYCSYSGYNDELL 265
>sptr|Q93WY8|Q93WY8 Endo-beta-1,4-glucanase (EC 3.2.1.4) (Fragment).
Length = 317
Score = 270 bits (689), Expect(2) = 3e-88
Identities = 127/229 (55%), Positives = 172/229 (75%)
Frame = -1
Query: 1039 AWLHKATRRREYREYIKRNEVVLGASDAINEFGWDNKHAGINVLISKEVLMGKDEYFQSF 860
AWLH+AT+ Y YI+RN +LGA++ N FGWDNKH G +L+SK L+ K + +
Sbjct: 90 AWLHRATKNPSYLSYIQRNGQILGAAETDNTFGWDNKHVGARILLSKAFLVQKLQSLHDY 149
Query: 859 RVNADNFMCTLLPGISNHPQIQYSPGGLLFKVGSSNMQHVTQLSFLLLAYSNYLSHAGGR 680
+ +ADN++C+L+PG + Q QY+PGGLLFK+ SNMQ+VT SFLL+ Y+ YL+ A
Sbjct: 150 KSHADNYICSLIPGTA-FSQAQYTPGGLLFKMDDSNMQYVTTTSFLLVTYAKYLTSARMV 208
Query: 679 VSCGSSSASPVQLRRVAKRQVDYILGDNPLRMSYMVGYGARFPRRIHHRASSLPSVAAHP 500
V CG +P +LR +AK+QVDY+LGDNPL+MS+MVGYGA +P+RIHHR SSLPSV+ HP
Sbjct: 209 VKCGGVVVTPKRLRNIAKKQVDYLLGDNPLKMSFMVGYGASYPQRIHHRGSSLPSVSNHP 268
Query: 499 ARIGCKAGAAYYTSPAPNPNLLVGAVVGGPTDDSDAFPDARAVFQQSEP 353
++I C++G + +S APNPN+LVGAVVGGP D+ D FPD R+ ++QSEP
Sbjct: 269 SQIECRSGFSVMSSQAPNPNVLVGAVVGGP-DEHDRFPDERSDYEQSEP 316
Score = 79.3 bits (194), Expect(2) = 3e-88
Identities = 35/56 (62%), Positives = 43/56 (76%)
Frame = -2
Query: 1218 SIVFRDADPAYSKRLLDRAVAVFEFADKYRGPYSSSLHAAVCPCYCDYSGYQDELL 1051
S+V+R +P+YSK L+ RA+ VFEFADKYRG YS+ L VCP YC SGY+DELL
Sbjct: 31 SLVYRKCNPSYSKLLVKRAIRVFEFADKYRGSYSNGLRKVVCPYYCSVSGYEDELL 86
>sptr|Q9AVI5|Q9AVI5 Endo-1,4-beta glucanase precursor.
Length = 494
Score = 263 bits (671), Expect(2) = 5e-88
Identities = 125/246 (50%), Positives = 169/246 (68%)
Frame = -1
Query: 1039 AWLHKATRRREYREYIKRNEVVLGASDAINEFGWDNKHAGINVLISKEVLMGKDEYFQSF 860
+W+H+A++ Y YI+ N +G+ D F WD+K G +L+SKE L E FQ +
Sbjct: 249 SWIHRASQNGSYLTYIQSNGHTMGSDDDDYSFSWDDKRPGTKILLSKEFLEKTTEEFQLY 308
Query: 859 RVNADNFMCTLLPGISNHPQIQYSPGGLLFKVGSSNMQHVTQLSFLLLAYSNYLSHAGGR 680
+ ++DN++C+L+PG S+ Q QY+PGGL +K SN+Q+VT +FLLL Y+ YL GG
Sbjct: 309 KSHSDNYICSLIPGTSSF-QAQYTPGGLFYKASESNLQYVTSTTFLLLTYAKYLGSNGGV 367
Query: 679 VSCGSSSASPVQLRRVAKRQVDYILGDNPLRMSYMVGYGARFPRRIHHRASSLPSVAAHP 500
CG S+ + L AK+QVDYILGDNP RMSYMVG+G R+P+ +HHR SS+PS+ AHP
Sbjct: 368 ARCGGSTVTTESLIAQAKKQVDYILGDNPARMSYMVGFGNRYPQHVHHRGSSVPSIHAHP 427
Query: 499 ARIGCKAGAAYYTSPAPNPNLLVGAVVGGPTDDSDAFPDARAVFQQSEPTTYINAPLMGL 320
RI C G + S +PNPN+LVGA++GGP D+ D F D R +QQSEP TYINAP +G
Sbjct: 428 NRISCNDGFQFLYSSSPNPNVLVGAIIGGP-DNRDNFADDRNNYQQSEPATYINAPFVGA 486
Query: 319 LAYFSA 302
LA+FSA
Sbjct: 487 LAFFSA 492
Score = 85.5 bits (210), Expect(2) = 5e-88
Identities = 37/56 (66%), Positives = 48/56 (85%)
Frame = -2
Query: 1218 SIVFRDADPAYSKRLLDRAVAVFEFADKYRGPYSSSLHAAVCPCYCDYSGYQDELL 1051
SIVF+++DP+YS +LL A+ VF+FAD+YRG YS+SL++ VCP YC YSGYQDELL
Sbjct: 190 SIVFKESDPSYSTKLLHTAMKVFDFADRYRGSYSNSLNSVVCPFYCSYSGYQDELL 245
>sptr|O22297|O22297 Acidic cellulase.
Length = 505
Score = 262 bits (670), Expect(2) = 1e-87
Identities = 126/246 (51%), Positives = 170/246 (69%)
Frame = -1
Query: 1039 AWLHKATRRREYREYIKRNEVVLGASDAINEFGWDNKHAGINVLISKEVLMGKDEYFQSF 860
+WLH+A++ Y YI+ N +LGA D F WD+K AG VL+SK L + FQ +
Sbjct: 257 SWLHRASQNSSYLAYIQSNGHILGADDDDYSFSWDDKRAGTKVLLSKGFLEKNTQEFQLY 316
Query: 859 RVNADNFMCTLLPGISNHPQIQYSPGGLLFKVGSSNMQHVTQLSFLLLAYSNYLSHAGGR 680
+ ++DN++C+L+PG S+ Q QY+ GGL +K SN+Q+VT +FLLL Y+ YLS GG
Sbjct: 317 KAHSDNYICSLIPGSSSF-QAQYTAGGLFYKASESNLQYVTTTAFLLLTYAKYLSSNGGV 375
Query: 679 VSCGSSSASPVQLRRVAKRQVDYILGDNPLRMSYMVGYGARFPRRIHHRASSLPSVAAHP 500
+CGSS+ L +AK+QVDYILGDNP +MSYMVG+G R+P+ +HHR SSLPS+ AHP
Sbjct: 376 ATCGSSTVKAENLIALAKKQVDYILGDNPAKMSYMVGFGERYPQHVHHRGSSLPSIHAHP 435
Query: 499 ARIGCKAGAAYYTSPAPNPNLLVGAVVGGPTDDSDAFPDARAVFQQSEPTTYINAPLMGL 320
I C G Y S +PNPN+L GA++GGP D+ D F D R +QQSEP TYINAP +G
Sbjct: 436 DHIACNDGFQYLYSRSPNPNVLTGAILGGP-DNRDNFADDRNNYQQSEPATYINAPFVGA 494
Query: 319 LAYFSA 302
+A+FS+
Sbjct: 495 VAFFSS 500
Score = 84.7 bits (208), Expect(2) = 1e-87
Identities = 37/56 (66%), Positives = 46/56 (82%)
Frame = -2
Query: 1218 SIVFRDADPAYSKRLLDRAVAVFEFADKYRGPYSSSLHAAVCPCYCDYSGYQDELL 1051
S+VF+D+DP+YS +LL A+ VF+FADKYRG YS SL++ VCP YC YSGY DELL
Sbjct: 198 SVVFKDSDPSYSTKLLKTAMKVFDFADKYRGSYSDSLNSVVCPYYCSYSGYLDELL 253
>sptr|Q9SML6|Q9SML6 Endo-beta-1,4-glucanase precursor (EC 3.2.1.4).
Length = 497
Score = 264 bits (675), Expect(2) = 2e-87
Identities = 129/246 (52%), Positives = 172/246 (69%)
Frame = -1
Query: 1039 AWLHKATRRREYREYIKRNEVVLGASDAINEFGWDNKHAGINVLISKEVLMGKDEYFQSF 860
+WLH+A++ Y YI+ N +GA+D F WD+K G +++SK+ L + FQ++
Sbjct: 249 SWLHRASQDTSYLSYIQSNGQTMGANDDDYSFSWDDKRPGTKIVLSKDFLEKSTQEFQAY 308
Query: 859 RVNADNFMCTLLPGISNHPQIQYSPGGLLFKVGSSNMQHVTQLSFLLLAYSNYLSHAGGR 680
+V++DN++C+L+PG + Q QY+PGGLLFK SN+Q+VT SFLLL Y+ YL GG
Sbjct: 309 KVHSDNYICSLIPGSPSF-QAQYTPGGLLFKGSESNLQYVTSSSFLLLTYAKYLRSNGGV 367
Query: 679 VSCGSSSASPVQLRRVAKRQVDYILGDNPLRMSYMVGYGARFPRRIHHRASSLPSVAAHP 500
VSCGSS +L +A++QVDYILGDNP ++SYMVG+G ++P R+HHR SSLPSV HP
Sbjct: 368 VSCGSSRFPANKLVELARKQVDYILGDNPAKISYMVGFGQKYPLRVHHRGSSLPSVRTHP 427
Query: 499 ARIGCKAGAAYYTSPAPNPNLLVGAVVGGPTDDSDAFPDARAVFQQSEPTTYINAPLMGL 320
IGC G S +PNPN+LVGA+VGGP D D F D R +QQSEP TYINAPL+G
Sbjct: 428 GHIGCNDGFQSLYSGSPNPNVLVGAIVGGP-DSRDNFEDDRNNYQQSEPATYINAPLVGA 486
Query: 319 LAYFSA 302
LA+ SA
Sbjct: 487 LAFLSA 492
Score = 82.0 bits (201), Expect(2) = 2e-87
Identities = 38/56 (67%), Positives = 42/56 (75%)
Frame = -2
Query: 1218 SIVFRDADPAYSKRLLDRAVAVFEFADKYRGPYSSSLHAAVCPCYCDYSGYQDELL 1051
SIVF+D+DP+YS LL A VF FADKYRG YS SL + VCP YC YSGY DELL
Sbjct: 190 SIVFKDSDPSYSSTLLHTAQKVFAFADKYRGSYSDSLSSVVCPFYCSYSGYNDELL 245
>sptr|Q96546|Q96546 Endo-beta-1,4-glucanase (EC 3.2.1.4).
Length = 497
Score = 264 bits (675), Expect(2) = 3e-87
Identities = 129/246 (52%), Positives = 172/246 (69%)
Frame = -1
Query: 1039 AWLHKATRRREYREYIKRNEVVLGASDAINEFGWDNKHAGINVLISKEVLMGKDEYFQSF 860
+WLH+A++ Y YI+ N +GA+D F WD+K G +++SK+ L + FQ++
Sbjct: 249 SWLHRASQDTSYLSYIQSNGQTMGANDDDYSFSWDDKRPGTKIVLSKDFLEKSTQEFQAY 308
Query: 859 RVNADNFMCTLLPGISNHPQIQYSPGGLLFKVGSSNMQHVTQLSFLLLAYSNYLSHAGGR 680
+V++DN++C+L+PG + Q QY+PGGLLFK SN+Q+VT SFLLL Y+ YL GG
Sbjct: 309 KVHSDNYICSLIPGSPSF-QAQYTPGGLLFKGSESNLQYVTSSSFLLLTYAKYLRSNGGV 367
Query: 679 VSCGSSSASPVQLRRVAKRQVDYILGDNPLRMSYMVGYGARFPRRIHHRASSLPSVAAHP 500
VSCGSS +L +A++QVDYILGDNP ++SYMVG+G ++P R+HHR SSLPSV HP
Sbjct: 368 VSCGSSRFPANKLVELARKQVDYILGDNPAKISYMVGFGQKYPLRVHHRGSSLPSVRTHP 427
Query: 499 ARIGCKAGAAYYTSPAPNPNLLVGAVVGGPTDDSDAFPDARAVFQQSEPTTYINAPLMGL 320
IGC G S +PNPN+LVGA+VGGP D D F D R +QQSEP TYINAPL+G
Sbjct: 428 GHIGCNDGFQSLYSGSPNPNVLVGAIVGGP-DSRDNFEDDRNNYQQSEPATYINAPLVGA 486
Query: 319 LAYFSA 302
LA+ SA
Sbjct: 487 LAFLSA 492
Score = 81.6 bits (200), Expect(2) = 3e-87
Identities = 38/56 (67%), Positives = 42/56 (75%)
Frame = -2
Query: 1218 SIVFRDADPAYSKRLLDRAVAVFEFADKYRGPYSSSLHAAVCPCYCDYSGYQDELL 1051
SIVF+D+DP+YS LL A VF FADKYRG YS SL + VCP YC YSGY DELL
Sbjct: 190 SIVFKDSDPSYSSTLLRTAQKVFAFADKYRGSYSDSLSSVVCPFYCSYSGYNDELL 245
>sptr|O82473|O82473 Endo-1,4-beta-glucanase (EC 3.2.1.4).
Length = 497
Score = 267 bits (682), Expect(2) = 5e-87
Identities = 131/249 (52%), Positives = 175/249 (70%)
Frame = -1
Query: 1039 AWLHKATRRREYREYIKRNEVVLGASDAINEFGWDNKHAGINVLISKEVLMGKDEYFQSF 860
+WLH+A++ Y YI+ N +GA+D F WD+K G +++SK+ L + FQ++
Sbjct: 252 SWLHRASQDSSYLAYIQSNGQTMGANDDDYSFSWDDKRPGTKIILSKDFLEKSSQEFQAY 311
Query: 859 RVNADNFMCTLLPGISNHPQIQYSPGGLLFKVGSSNMQHVTQLSFLLLAYSNYLSHAGGR 680
+V++DN++C+L+PG + Q QY+PGGLL+K SN+Q+VT SFLLL Y+ YL GG
Sbjct: 312 KVHSDNYICSLIPGSPSF-QAQYTPGGLLYKGSGSNLQYVTSSSFLLLTYAKYLRSNGGD 370
Query: 679 VSCGSSSASPVQLRRVAKRQVDYILGDNPLRMSYMVGYGARFPRRIHHRASSLPSVAAHP 500
VSCGSS +L +AK+QVDYILGDNP ++SYMVG+G ++P R+HHR SSLPSV AHP
Sbjct: 371 VSCGSSRFPAERLVELAKKQVDYILGDNPAKISYMVGFGDKYPLRVHHRGSSLPSVHAHP 430
Query: 499 ARIGCKAGAAYYTSPAPNPNLLVGAVVGGPTDDSDAFPDARAVFQQSEPTTYINAPLMGL 320
IGC G S +PNPN+L+GA+VGGP D D F D R +QQSEP TYINAPL+G
Sbjct: 431 GHIGCNDGFQSLNSGSPNPNVLIGAIVGGP-DSRDNFEDDRNNYQQSEPATYINAPLVGA 489
Query: 319 LAYFSAHPN 293
LA+ SA PN
Sbjct: 490 LAFLSA-PN 497
Score = 78.2 bits (191), Expect(2) = 5e-87
Identities = 37/56 (66%), Positives = 41/56 (73%)
Frame = -2
Query: 1218 SIVFRDADPAYSKRLLDRAVAVFEFADKYRGPYSSSLHAAVCPCYCDYSGYQDELL 1051
SIVF+D+DP+YS LL A VF FADKYRG YS SL + VCP C YSGY DELL
Sbjct: 193 SIVFKDSDPSYSSSLLRTAQKVFAFADKYRGSYSDSLSSVVCPFSCSYSGYNDELL 248
Database: /db/uniprot/tmp/swall
Posted date: Mar 5, 2004 7:45 PM
Number of letters in database: 442,889,342
Number of sequences in database: 1,395,590
Lambda K H
0.318 0.135 0.401
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 744,134,718
Number of Sequences: 1395590
Number of extensions: 16403134
Number of successful extensions: 52036
Number of sequences better than 10.0: 30
Number of HSP's better than 10.0 without gapping: 46989
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 51442
length of database: 442,889,342
effective HSP length: 126
effective length of database: 267,045,002
effective search space used: 74772600560
frameshift window, decay const: 50, 0.1
T: 12
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)

