BLASTX 2.2.6 [Apr-09-2003]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= 2493528.2.1
(627 letters)
Database: /db/trembl-ebi/tmp/swall
1,272,877 sequences; 406,886,216 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
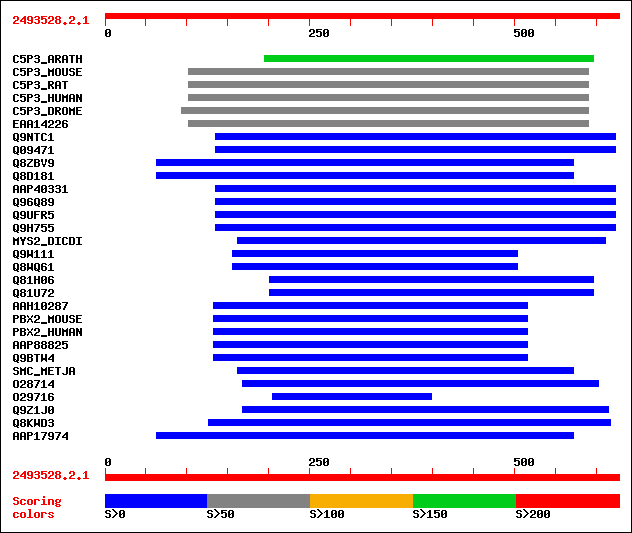
sw|Q9FG23|C5P3_ARATH CDK5RAP3-like protein. 190 1e-47
sw|Q99LM2|C5P3_MOUSE CDK5 regulatory subunit associated protein ... 79 3e-14
sw|Q9JLH7|C5P3_RAT CDK5 regulatory subunit associated protein 3 ... 79 6e-14
sw|Q96JB5|C5P3_HUMAN CDK5 regulatory subunit associated protein ... 78 8e-14
sw|Q95SK3|C5P3_DROME CDK5RAP3-like protein. 62 7e-09
sptrnew|EAA14226|EAA14226 AgCP8594 (Fragment). 57 1e-07
sptr|Q9NTC1|Q9NTC1 Hypothetical protein (Fragment). 39 0.051
sptr|Q09471|Q09471 M-phase phosphoprotein-1 (MPP1) (Fragment). 38 0.087
sptr|Q8ZBV9|Q8ZBV9 Clp ATPase. 38 0.087
sptr|Q8D181|Q8D181 Heat shock protein. 38 0.087
sptrnew|AAP40331|AAP40331 M-phase phosphoprotein 1 (Fragment). 38 0.087
sptr|Q96Q89|Q96Q89 Mitotic kinesin-related protein. 38 0.087
sptr|Q9UFR5|Q9UFR5 Hypothetical protein. 38 0.087
sptr|Q9H755|Q9H755 Hypothetical protein FLJ21306 (Fragment). 37 0.19
sw|P08799|MYS2_DICDI Myosin II heavy chain, non muscle. 37 0.19
sptr|Q9W111|Q9W111 CG16932 protein. 35 0.57
sptr|Q8WQ61|Q8WQ61 Eps-15 protein (SD09478p). 35 0.57
sptr|Q81H06|Q81H06 CMP-binding factor. 34 1.3
sptr|Q81U72|Q81U72 HD domain protein. 34 1.3
sptrnew|AAH10287|AAH10287 Pre B-cell leukemia transcription fact... 34 1.6
sw|O35984|PBX2_MOUSE Pre-B-cell leukemia transcription factor-2 ... 34 1.6
sw|P40425|PBX2_HUMAN Pre-B-cell leukemia transcription factor-2 ... 34 1.6
sptrnew|AAP88825|AAP88825 Pre-B-cell leukemia transcription fact... 34 1.6
sptr|Q9BTW4|Q9BTW4 Similar to pre-B-cell leukemia transcription ... 34 1.6
sw|Q59037|SMC_METJA Chromosome partition protein smc homolog. 33 2.1
sptr|O28714|O28714 Chromosome segregation protein (SMC1). 33 2.1
sptr|O29716|O29716 Hypothetical protein AF0534. 33 2.1
sptr|Q9Z1J0|Q9Z1J0 Kinesin-related mitotic motor protein (Fragme... 33 2.8
sptr|Q8KWD3|Q8KWD3 RB109. 33 3.7
sptrnew|AAP17974|AAP17974 Heat shock protein. 33 3.7
>sw|Q9FG23|C5P3_ARATH CDK5RAP3-like protein.
Length = 522
Score = 190 bits (482), Expect = 1e-47
Identities = 98/134 (73%), Positives = 114/134 (85%)
Frame = -2
Query: 596 DRSLLLEKEYRNNILDDLLEVKSFLTQRLGEMRNADTSSLQHQVQAVSPFVLQQHAPDSL 417
+RS LLE EYRN ILDDL EVK+FL QRL E+RN DT SLQH VQAVSP VLQQ++P+++
Sbjct: 384 ERSQLLETEYRNKILDDLYEVKAFLNQRLIELRNEDTLSLQHHVQAVSPMVLQQYSPETI 443
Query: 416 ENMLVEISSAISLLTNQKTLDLIMILNSKRFLDRLVSSLEEKKHHEVKLREGLGDLSVKR 237
E M+V+IS AISLLTN+K+ DLIMILNSKRFLDRLVS LEEKKH EVKLRE L D+ +R
Sbjct: 444 EPMVVDISMAISLLTNKKSRDLIMILNSKRFLDRLVSELEEKKHREVKLRESLKDVGRRR 503
Query: 236 MELQNALSSSWPKQ 195
MELQN+LS+ WPKQ
Sbjct: 504 MELQNSLSAIWPKQ 517
>sw|Q99LM2|C5P3_MOUSE CDK5 regulatory subunit associated protein 3
(CDK5 activator-binding protein C53).
Length = 503
Score = 79.3 bits (194), Expect = 3e-14
Identities = 54/164 (32%), Positives = 90/164 (54%), Gaps = 1/164 (0%)
Frame = -2
Query: 590 SLLLEKEYRNNILDDLLEVKSFLTQRLGEM-RNADTSSLQHQVQAVSPFVLQQHAPDSLE 414
+LL E RN +D+L+E++ FL+QR EM AD S+ Q Q ++P +LQ + +
Sbjct: 339 TLLEYPETRNQFIDELMELEIFLSQRAVEMSEEADILSVS-QFQ-LAPAILQGQTKEKML 396
Query: 413 NMLVEISSAISLLTNQKTLDLIMILNSKRFLDRLVSSLEEKKHHEVKLREGLGDLSVKRM 234
+++ + I LT+ + L MIL S R++DR+ L++K L + K+
Sbjct: 397 SLVSTLQQLIGRLTSLRMQHLFMILASPRYVDRVTEFLQQKLKQSQLLALKKELMVEKQQ 456
Query: 233 ELQNALSSSWPKQEAAITKTRELKKLCESTLSSVFDGRPVYIIG 102
E ++ PK + + KTREL+KL E+ +S + GRPV ++G
Sbjct: 457 EALQEQAALEPKLDLLLEKTRELQKLIEADISKRYSGRPVNLMG 500
>sw|Q9JLH7|C5P3_RAT CDK5 regulatory subunit associated protein 3
(CDK5 activator-binding protein C53).
Length = 504
Score = 78.6 bits (192), Expect = 6e-14
Identities = 54/164 (32%), Positives = 90/164 (54%), Gaps = 1/164 (0%)
Frame = -2
Query: 590 SLLLEKEYRNNILDDLLEVKSFLTQRLGEM-RNADTSSLQHQVQAVSPFVLQQHAPDSLE 414
+LL E RN +D+L+E++ FL+QR EM AD S+ Q Q ++P +LQ + +
Sbjct: 340 TLLEYPETRNQFIDELMELEIFLSQRAVEMSEEADILSVS-QFQ-LAPAILQGQTKEKML 397
Query: 413 NMLVEISSAISLLTNQKTLDLIMILNSKRFLDRLVSSLEEKKHHEVKLREGLGDLSVKRM 234
+++ + I LT+ L MIL S R++DR+ L++K L + K+
Sbjct: 398 SLVSTLQHLIGQLTSLDLQHLFMILASPRYVDRVTELLQQKLKQSQLLALKKDLMVEKQQ 457
Query: 233 ELQNALSSSWPKQEAAITKTRELKKLCESTLSSVFDGRPVYIIG 102
E ++ PK + + KTREL+KL E+ +S ++GRPV ++G
Sbjct: 458 EALQEQAALEPKLDLLLEKTRELQKLIEADISKRYNGRPVNLMG 501
>sw|Q96JB5|C5P3_HUMAN CDK5 regulatory subunit associated protein 3
(CDK5 activator-binding protein C53) (HSF-27 protein)
(MSTP016) (PP1553).
Length = 506
Score = 78.2 bits (191), Expect = 8e-14
Identities = 54/164 (32%), Positives = 88/164 (53%), Gaps = 1/164 (0%)
Frame = -2
Query: 590 SLLLEKEYRNNILDDLLEVKSFLTQRLGEM-RNADTSSLQHQVQAVSPFVLQQHAPDSLE 414
+LL E RN LD+L+E++ FL QR E+ AD S+ Q Q ++P +LQ + +
Sbjct: 342 TLLEYTETRNQFLDELMELEIFLAQRAVELSEEADVLSVS-QFQ-LAPAILQGQTKEKMV 399
Query: 413 NMLVEISSAISLLTNQKTLDLIMILNSKRFLDRLVSSLEEKKHHEVKLREGLGDLSVKRM 234
M+ + I LT+ + L MIL S R++DR+ L++K L + K+
Sbjct: 400 TMVSVLEDLIGKLTSLQLQHLFMILASPRYVDRVTEFLQQKLKQSQLLALKKELMVQKQQ 459
Query: 233 ELQNALSSSWPKQEAAITKTRELKKLCESTLSSVFDGRPVYIIG 102
E ++ PK + + KT+EL+KL E+ +S + GRPV ++G
Sbjct: 460 EALEEQAALEPKLDLLLEKTKELQKLIEADISKRYSGRPVNLMG 503
>sw|Q95SK3|C5P3_DROME CDK5RAP3-like protein.
Length = 509
Score = 61.6 bits (148), Expect = 7e-09
Identities = 42/166 (25%), Positives = 80/166 (48%)
Frame = -2
Query: 590 SLLLEKEYRNNILDDLLEVKSFLTQRLGEMRNADTSSLQHQVQAVSPFVLQQHAPDSLEN 411
+LL YR+ LD++ E++SFL R+ E++ ++SS + + H +S+
Sbjct: 347 TLLDSPNYRDRFLDEIYELESFLRMRIYELKQLESSS---DIMFSLMDNIATHDGESIWK 403
Query: 410 MLVEISSAISLLTNQKTLDLIMILNSKRFLDRLVSSLEEKKHHEVKLREGLGDLSVKRME 231
+LV + I ++++T L + +S ++ + L + L++ KLR L +E
Sbjct: 404 ILVSVEKIIQQTSDKQTQHLFQLKHSPKYANMLATKLQQMTKAVEKLRATREALKQLTIE 463
Query: 230 LQNALSSSWPKQEAAITKTRELKKLCESTLSSVFDGRPVYIIGEIN 93
L+ P E I +TR L+ E +S + R V ++G +N
Sbjct: 464 LREQRQDLNPVLEELIAQTRTLQSHIEKDISKRYKNRVVNLMGGVN 509
>sptrnew|EAA14226|EAA14226 AgCP8594 (Fragment).
Length = 535
Score = 57.4 bits (137), Expect = 1e-07
Identities = 42/164 (25%), Positives = 84/164 (51%), Gaps = 1/164 (0%)
Frame = -2
Query: 590 SLLLEKEYRNNILDDLLEVKSFLTQRLGEMRNADTSSLQHQVQAVSPF-VLQQHAPDSLE 414
++L ++YR+ ++DLLE++SFL RL ++ +AD + QV A + + H ++
Sbjct: 373 TVLDAQQYRDRFVNDLLELQSFLKMRLHQLGSADKA----QVLAFAIMDNFKDHDEKTVA 428
Query: 413 NMLVEISSAISLLTNQKTLDLIMILNSKRFLDRLVSSLEEKKHHEVKLREGLGDLSVKRM 234
+ML E+ + +T++ L+ I +S ++D L S++++K K++E K +
Sbjct: 429 SMLAEVEVVYAGVTDELLQHLLQIKHSPVYVDILTSNVKQKLTSIDKMKETALMKREKSV 488
Query: 233 ELQNALSSSWPKQEAAITKTRELKKLCESTLSSVFDGRPVYIIG 102
+ P Q + +T+ L+ E +S + RPV + G
Sbjct: 489 SFKQQSVELKPTQTKIVEQTKTLQGQIEKDISKRYKNRPVNLYG 532
>sptr|Q9NTC1|Q9NTC1 Hypothetical protein (Fragment).
Length = 1206
Score = 38.9 bits (89), Expect = 0.051
Identities = 47/184 (25%), Positives = 83/184 (45%), Gaps = 21/184 (11%)
Frame = -2
Query: 623 RAQSQMIAEDRSLLLEKEYRNNILDDLLEVKSFLTQRLGEMRNADTSSLQHQVQ------ 462
+A+ + ++ + L EKE++N DDLL+ K L Q+L E +L Q+Q
Sbjct: 475 QAEVKGYKDENNRLKEKEHKNQ--DDLLKEKETLIQQLKEELQEKNVTLDVQIQHVVEGK 532
Query: 461 -AVSPFVLQ--------QHAPDSLENMLVEISSAISL---LTNQKTLDLIMILNSKRFLD 318
A+S + LE VE S + L + ++++ L + N K F +
Sbjct: 533 RALSELTQGVTCYKAKIKELETILETQKVECSHSAKLEQDILEKESIILKLERNLKEFQE 592
Query: 317 RLVSSLEEKKH---HEVKLREGLGDLSVKRMELQNALSSSWPKQEAAITKTRELKKLCES 147
L S++ K E+KL+E + L+ ++++ L K+E T +E +KL E
Sbjct: 593 HLQDSVKNTKDLNVKELKLKEEITQLTNNLQDMKHLLQL---KEEEEETNRQETEKLKEE 649
Query: 146 TLSS 135
+S
Sbjct: 650 LSAS 653
>sptr|Q09471|Q09471 M-phase phosphoprotein-1 (MPP1) (Fragment).
Length = 753
Score = 38.1 bits (87), Expect = 0.087
Identities = 47/184 (25%), Positives = 83/184 (45%), Gaps = 21/184 (11%)
Frame = -2
Query: 623 RAQSQMIAEDRSLLLEKEYRNNILDDLLEVKSFLTQRLGEMRNADTSSLQHQVQ------ 462
+A+ + ++ + L EKE++N DDLL+ K L Q+L E +L Q+Q
Sbjct: 22 QAEVKGYKDENNRLKEKEHKNQ--DDLLKEKETLIQQLKEELQEKNVTLDVQIQHVVEGK 79
Query: 461 -AVSPFVLQ--------QHAPDSLENMLVEISSAISL---LTNQKTLDLIMILNSKRFLD 318
A+S + LE VE S + L + ++++ L + N K F +
Sbjct: 80 RALSELTQGVTCYKAKIKELETILETQKVERSHSAKLEQDILEKESIILKLERNLKEFQE 139
Query: 317 RLVSSLEEKKH---HEVKLREGLGDLSVKRMELQNALSSSWPKQEAAITKTRELKKLCES 147
L S++ K E+KL+E + L+ ++++ L K+E T +E +KL E
Sbjct: 140 HLQDSVKNTKDLNVKELKLKEEITQLTNNLQDMKHLLQL---KEEEEETNRQETEKLKEE 196
Query: 146 TLSS 135
+S
Sbjct: 197 LSAS 200
>sptr|Q8ZBV9|Q8ZBV9 Clp ATPase.
Length = 857
Score = 38.1 bits (87), Expect = 0.087
Identities = 43/202 (21%), Positives = 76/202 (37%), Gaps = 32/202 (15%)
Frame = -2
Query: 572 EYRNNILDDLLEVKSFLTQRLGEMRNADTSS----------LQHQVQAVSP--------- 450
EYR I D + F + E DT + L H VQ P
Sbjct: 318 EYRQYIEKDAALERRFQKVYVAEPTVEDTIAILRGLKERYELHHHVQITDPAIVAAATLS 377
Query: 449 --FVLQQHAPDSLENMLVEISSAISLLTNQKTLDLIMILNSKRFLDRLVSSLEEKKHHEV 276
++ + PD +++ E S+I + + K LDRL + + K +
Sbjct: 378 HRYISDRQLPDKAIDLIDEAGSSIRMQMDSKP----------ESLDRLERRIIQLKLEQQ 427
Query: 275 KLREGLGDLSVKRMELQNA-----------LSSSWPKQEAAITKTRELKKLCESTLSSVF 129
L++ D S KR+E+ N L W ++A++T T+ +K E ++
Sbjct: 428 ALKKESDDASKKRLEMLNTELEQKEREYSELEEEWKAEKASLTGTQNIKTELEQAKITLE 487
Query: 128 DGRPVYIIGEINTLLSSSVSQL 63
R V + ++ L ++ +L
Sbjct: 488 QARRVGDLARMSELQYGTIPEL 509
>sptr|Q8D181|Q8D181 Heat shock protein.
Length = 864
Score = 38.1 bits (87), Expect = 0.087
Identities = 43/202 (21%), Positives = 76/202 (37%), Gaps = 32/202 (15%)
Frame = -2
Query: 572 EYRNNILDDLLEVKSFLTQRLGEMRNADTSS----------LQHQVQAVSP--------- 450
EYR I D + F + E DT + L H VQ P
Sbjct: 325 EYRQYIEKDAALERRFQKVYVAEPTVEDTIAILRGLKERYELHHHVQITDPAIVAAATLS 384
Query: 449 --FVLQQHAPDSLENMLVEISSAISLLTNQKTLDLIMILNSKRFLDRLVSSLEEKKHHEV 276
++ + PD +++ E S+I + + K LDRL + + K +
Sbjct: 385 HRYISDRQLPDKAIDLIDEAGSSIRMQMDSKP----------ESLDRLERRIIQLKLEQQ 434
Query: 275 KLREGLGDLSVKRMELQNA-----------LSSSWPKQEAAITKTRELKKLCESTLSSVF 129
L++ D S KR+E+ N L W ++A++T T+ +K E ++
Sbjct: 435 ALKKESDDASKKRLEMLNTELEQKEREYSELEEEWKAEKASLTGTQNIKTELEQAKITLE 494
Query: 128 DGRPVYIIGEINTLLSSSVSQL 63
R V + ++ L ++ +L
Sbjct: 495 QARRVGDLARMSELQYGTIPEL 516
>sptrnew|AAP40331|AAP40331 M-phase phosphoprotein 1 (Fragment).
Length = 1116
Score = 38.1 bits (87), Expect = 0.087
Identities = 47/184 (25%), Positives = 83/184 (45%), Gaps = 21/184 (11%)
Frame = -2
Query: 623 RAQSQMIAEDRSLLLEKEYRNNILDDLLEVKSFLTQRLGEMRNADTSSLQHQVQ------ 462
+A+ + ++ + L EKE++N DDLL+ K L Q+L E +L Q+Q
Sbjct: 385 QAEVKGYKDENNRLKEKEHKNQ--DDLLKEKETLIQQLKEELQEKNVTLDVQIQHVVEGK 442
Query: 461 -AVSPFVLQ--------QHAPDSLENMLVEISSAISL---LTNQKTLDLIMILNSKRFLD 318
A+S + LE VE S + L + ++++ L + N K F +
Sbjct: 443 RALSELTQGVTCYKAKIKELETILETQKVERSHSAKLEQDILEKESIILKLERNLKEFQE 502
Query: 317 RLVSSLEEKKH---HEVKLREGLGDLSVKRMELQNALSSSWPKQEAAITKTRELKKLCES 147
L S++ K E+KL+E + L+ ++++ L K+E T +E +KL E
Sbjct: 503 HLQDSVKNTKDLNVKELKLKEEITQLTNNLQDMKHLLQL---KEEEEETNRQETEKLKEE 559
Query: 146 TLSS 135
+S
Sbjct: 560 LSAS 563
>sptr|Q96Q89|Q96Q89 Mitotic kinesin-related protein.
Length = 1820
Score = 38.1 bits (87), Expect = 0.087
Identities = 47/184 (25%), Positives = 83/184 (45%), Gaps = 21/184 (11%)
Frame = -2
Query: 623 RAQSQMIAEDRSLLLEKEYRNNILDDLLEVKSFLTQRLGEMRNADTSSLQHQVQ------ 462
+A+ + ++ + L EKE++N DDLL+ K L Q+L E +L Q+Q
Sbjct: 1089 QAEVKGYKDENNRLKEKEHKNQ--DDLLKEKETLIQQLKEELQEKNVTLDVQIQHVVEGK 1146
Query: 461 -AVSPFVLQ--------QHAPDSLENMLVEISSAISL---LTNQKTLDLIMILNSKRFLD 318
A+S + LE VE S + L + ++++ L + N K F +
Sbjct: 1147 RALSELTQGVTCYKAKIKELETILETQKVERSHSAKLEQDILEKESIILKLERNLKEFQE 1206
Query: 317 RLVSSLEEKKH---HEVKLREGLGDLSVKRMELQNALSSSWPKQEAAITKTRELKKLCES 147
L S++ K E+KL+E + L+ ++++ L K+E T +E +KL E
Sbjct: 1207 HLQDSVKNTKDLNVKELKLKEEITQLTNNLQDMKHLLQL---KEEEEETNRQETEKLKEE 1263
Query: 146 TLSS 135
+S
Sbjct: 1264 LSAS 1267
>sptr|Q9UFR5|Q9UFR5 Hypothetical protein.
Length = 1780
Score = 38.1 bits (87), Expect = 0.087
Identities = 47/184 (25%), Positives = 83/184 (45%), Gaps = 21/184 (11%)
Frame = -2
Query: 623 RAQSQMIAEDRSLLLEKEYRNNILDDLLEVKSFLTQRLGEMRNADTSSLQHQVQ------ 462
+A+ + ++ + L EKE++N DDLL+ K L Q+L E +L Q+Q
Sbjct: 1049 QAEVKGYKDENNRLKEKEHKNQ--DDLLKEKETLIQQLKEELQEKNVTLDVQIQHVVEGK 1106
Query: 461 -AVSPFVLQ--------QHAPDSLENMLVEISSAISL---LTNQKTLDLIMILNSKRFLD 318
A+S + LE VE S + L + ++++ L + N K F +
Sbjct: 1107 RALSELTQGVTCYKAKIKELETILETQKVERSHSAKLEQDILEKESIILKLERNLKEFQE 1166
Query: 317 RLVSSLEEKKH---HEVKLREGLGDLSVKRMELQNALSSSWPKQEAAITKTRELKKLCES 147
L S++ K E+KL+E + L+ ++++ L K+E T +E +KL E
Sbjct: 1167 HLQDSVKNTKDLNVKELKLKEEITQLTNNLQDMKHLLQL---KEEEEETNRQETEKLKEE 1223
Query: 146 TLSS 135
+S
Sbjct: 1224 LSAS 1227
>sptr|Q9H755|Q9H755 Hypothetical protein FLJ21306 (Fragment).
Length = 696
Score = 37.0 bits (84), Expect = 0.19
Identities = 46/184 (25%), Positives = 83/184 (45%), Gaps = 21/184 (11%)
Frame = -2
Query: 623 RAQSQMIAEDRSLLLEKEYRNNILDDLLEVKSFLTQRLGEMRNADTSSLQHQVQ------ 462
+A+ + ++ + L E+E++N DDLL+ K L Q+L E +L Q+Q
Sbjct: 300 QAEVKGYKDENNRLKEREHKNQ--DDLLKEKETLIQQLKEELQEKNVTLDVQIQHVVEGK 357
Query: 461 -AVSPFVLQ--------QHAPDSLENMLVEISSAISL---LTNQKTLDLIMILNSKRFLD 318
A+S + LE VE S + L + ++++ L + N K F +
Sbjct: 358 RALSELTQGVTCYKAKIKELETILETQKVERSHSAKLEQDILEKESIILKLERNLKEFQE 417
Query: 317 RLVSSLEEKKH---HEVKLREGLGDLSVKRMELQNALSSSWPKQEAAITKTRELKKLCES 147
L S++ K E+KL+E + L+ ++++ L K+E T +E +KL E
Sbjct: 418 HLQDSVKNTKDLNVKELKLKEEITQLTNNLQDMKHLLQL---KEEEEETNRQETEKLKEE 474
Query: 146 TLSS 135
+S
Sbjct: 475 LSAS 478
>sw|P08799|MYS2_DICDI Myosin II heavy chain, non muscle.
Length = 2116
Score = 37.0 bits (84), Expect = 0.19
Identities = 40/166 (24%), Positives = 72/166 (43%), Gaps = 16/166 (9%)
Frame = -2
Query: 611 QMIAEDRSLLLEKEYRNNILD---DLLEVKSFLTQRLGEMRNADTS---------SLQHQ 468
++ ++ R LL + + I + ++LE+KS LT + + S LQ Q
Sbjct: 812 KLFSKARPLLKRRNFEKEIKEKEREILELKSNLTDSTTQKDKLEKSLKDTESNVLDLQRQ 871
Query: 467 VQAVSPFVLQQH-APDSLENMLVEISSAISLLTNQ---KTLDLIMILNSKRFLDRLVSSL 300
++A + + + D+LE E+ + + ++ K L L + N KR ++ V L
Sbjct: 872 LKAEKETLKAMYDSKDALEAQKRELEIRVEDMESELDEKKLALENLQNQKRSVEEKVRDL 931
Query: 299 EEKKHHEVKLREGLGDLSVKRMELQNALSSSWPKQEAAITKTRELK 162
EE+ E KLR L L K E + Q I++ ++K
Sbjct: 932 EEELQEEQKLRNTLEKLKKKYEEELEEMKRVNDGQSDTISRLEKIK 977
>sptr|Q9W111|Q9W111 CG16932 protein.
Length = 1253
Score = 35.4 bits (80), Expect = 0.57
Identities = 31/120 (25%), Positives = 54/120 (45%), Gaps = 4/120 (3%)
Frame = -2
Query: 503 MRNADTSSLQHQVQAVSPFVLQQHAPDSLENMLVEISSAISLLTNQKTLDLIMILNSKRF 324
++N + SLQ ++ ++ + Q LEN E + L Q + + ++ N
Sbjct: 454 IKNGEVRSLQSELDTLTATLKQ------LENQRGEAQKRLDDLQAQVSHNTAVLANVSLD 507
Query: 323 LDRL---VSSLEEKKH-HEVKLREGLGDLSVKRMELQNALSSSWPKQEAAITKTRELKKL 156
+ R V+ + ++ H EV + E G+L+ KR ELQ Q+ + REL KL
Sbjct: 508 ISRTNEQVTKIRDQCHMQEVTINEQEGELNAKRSELQKLKDEEASLQKEYDSNNRELSKL 567
>sptr|Q8WQ61|Q8WQ61 Eps-15 protein (SD09478p).
Length = 1253
Score = 35.4 bits (80), Expect = 0.57
Identities = 31/120 (25%), Positives = 54/120 (45%), Gaps = 4/120 (3%)
Frame = -2
Query: 503 MRNADTSSLQHQVQAVSPFVLQQHAPDSLENMLVEISSAISLLTNQKTLDLIMILNSKRF 324
++N + SLQ ++ ++ + Q LEN E + L Q + + ++ N
Sbjct: 454 IKNGEVRSLQSELDTLTATLKQ------LENQRGEAQKRLDDLQAQVSHNTAVLANVSLD 507
Query: 323 LDRL---VSSLEEKKH-HEVKLREGLGDLSVKRMELQNALSSSWPKQEAAITKTRELKKL 156
+ R V+ + ++ H EV + E G+L+ KR ELQ Q+ + REL KL
Sbjct: 508 ISRTNEQVTKIRDQCHMQEVTINEQEGELNAKRSELQKLKDEEASLQKEYDSNNRELSKL 567
>sptr|Q81H06|Q81H06 CMP-binding factor.
Length = 314
Score = 34.3 bits (77), Expect = 1.3
Identities = 37/136 (27%), Positives = 63/136 (46%), Gaps = 4/136 (2%)
Frame = -2
Query: 596 DRSLLLEKEYRNN----ILDDLLEVKSFLTQRLGEMRNADTSSLQHQVQAVSPFVLQQHA 429
D S +EK+Y + D+L K + R+ ++R A+ +++V +S FV + A
Sbjct: 52 DVSPEVEKQYVAETIVKVAGDILNYKGRIQLRVKQIRVAN----ENEVTDISDFV--EKA 105
Query: 428 PDSLENMLVEISSAISLLTNQKTLDLIMILNSKRFLDRLVSSLEEKKHHEVKLREGLGDL 249
P E+M+ +I+ I + N L L +K + L K HHE GL
Sbjct: 106 PVKKEDMVEKITQYIFEMRNPNIQRLTRHLLNKHQNEFLDYPAATKNHHE--FVSGLAYH 163
Query: 248 SVKRMELQNALSSSWP 201
V ++L A+S+ +P
Sbjct: 164 VVSMLDLAKAISNLYP 179
>sptr|Q81U72|Q81U72 HD domain protein.
Length = 314
Score = 34.3 bits (77), Expect = 1.3
Identities = 37/136 (27%), Positives = 63/136 (46%), Gaps = 4/136 (2%)
Frame = -2
Query: 596 DRSLLLEKEYRNN----ILDDLLEVKSFLTQRLGEMRNADTSSLQHQVQAVSPFVLQQHA 429
D S +EK+Y + D+L K + R+ ++R A+ +++V +S FV + A
Sbjct: 52 DVSPEVEKQYVAETIVKVAGDILNYKGRIQLRVKQIRVAN----ENEVTDISDFV--EKA 105
Query: 428 PDSLENMLVEISSAISLLTNQKTLDLIMILNSKRFLDRLVSSLEEKKHHEVKLREGLGDL 249
P E+M+ +I+ I + N L L +K + L K HHE GL
Sbjct: 106 PVKKEDMVEKITQYIFEMRNPNIQRLTRHLLNKHQNEFLDYPAATKNHHE--FVSGLAYH 163
Query: 248 SVKRMELQNALSSSWP 201
V ++L A+S+ +P
Sbjct: 164 VVSMLDLAKAISNLYP 179
>sptrnew|AAH10287|AAH10287 Pre B-cell leukemia transcription factor
2.
Length = 430
Score = 33.9 bits (76), Expect = 1.6
Identities = 40/143 (27%), Positives = 66/143 (46%), Gaps = 15/143 (10%)
Frame = -2
Query: 515 RLGEMRNADTSSLQHQVQAVSPF------VLQQH------APDSLENMLVEIS---SAIS 381
+L ++R+ S L+ QA + F +L++ AP +E M+ I SAI
Sbjct: 164 KLAQIRHIYHSELEKYEQACNEFTTHVMNLLREQSRTRPVAPKEMERMVSIIHRKFSAIQ 223
Query: 380 LLTNQKTLDLIMILNSKRFLDRLVSSLEEKKHHEVKLREGLGDLSVKRMELQNALSSSWP 201
+ Q T + +MIL S RFLD + ++++ + E L + + LS+ +P
Sbjct: 224 MQLKQSTCEAVMILRS-RFLD----ARRKRRNFSKQATEVLNEY------FYSHLSNPYP 272
Query: 200 KQEAAITKTRELKKLCESTLSSV 132
+EA EL K C T+S V
Sbjct: 273 SEEA----KEELAKKCGITVSQV 291
>sw|O35984|PBX2_MOUSE Pre-B-cell leukemia transcription factor-2
(Homeobox protein PBX2).
Length = 430
Score = 33.9 bits (76), Expect = 1.6
Identities = 40/143 (27%), Positives = 66/143 (46%), Gaps = 15/143 (10%)
Frame = -2
Query: 515 RLGEMRNADTSSLQHQVQAVSPF------VLQQH------APDSLENMLVEIS---SAIS 381
+L ++R+ S L+ QA + F +L++ AP +E M+ I SAI
Sbjct: 164 KLAQIRHIYHSELEKYEQACNEFTTHVMNLLREQSRTRPVAPKEMERMVSIIHRKFSAIQ 223
Query: 380 LLTNQKTLDLIMILNSKRFLDRLVSSLEEKKHHEVKLREGLGDLSVKRMELQNALSSSWP 201
+ Q T + +MIL S RFLD + ++++ + E L + + LS+ +P
Sbjct: 224 MQLKQSTCEAVMILRS-RFLD----ARRKRRNFSKQATEVLNEY------FYSHLSNPYP 272
Query: 200 KQEAAITKTRELKKLCESTLSSV 132
+EA EL K C T+S V
Sbjct: 273 SEEA----KEELAKKCGITVSQV 291
>sw|P40425|PBX2_HUMAN Pre-B-cell leukemia transcription factor-2
(Homeobox protein PBX2) (G17 protein).
Length = 430
Score = 33.9 bits (76), Expect = 1.6
Identities = 40/143 (27%), Positives = 66/143 (46%), Gaps = 15/143 (10%)
Frame = -2
Query: 515 RLGEMRNADTSSLQHQVQAVSPF------VLQQH------APDSLENMLVEIS---SAIS 381
+L ++R+ S L+ QA + F +L++ AP +E M+ I SAI
Sbjct: 164 KLAQIRHIYHSELEKYEQACNEFTTHVMNLLREQSRTRPVAPKEMERMVSIIHRKFSAIQ 223
Query: 380 LLTNQKTLDLIMILNSKRFLDRLVSSLEEKKHHEVKLREGLGDLSVKRMELQNALSSSWP 201
+ Q T + +MIL S RFLD + ++++ + E L + + LS+ +P
Sbjct: 224 MQLKQSTCEAVMILRS-RFLD----ARRKRRNFSKQATEVLNEY------FYSHLSNPYP 272
Query: 200 KQEAAITKTRELKKLCESTLSSV 132
+EA EL K C T+S V
Sbjct: 273 SEEA----KEELAKKCGITVSQV 291
>sptrnew|AAP88825|AAP88825 Pre-B-cell leukemia transcription factor
2.
Length = 349
Score = 33.9 bits (76), Expect = 1.6
Identities = 40/143 (27%), Positives = 66/143 (46%), Gaps = 15/143 (10%)
Frame = -2
Query: 515 RLGEMRNADTSSLQHQVQAVSPF------VLQQH------APDSLENMLVEIS---SAIS 381
+L ++R+ S L+ QA + F +L++ AP +E M+ I SAI
Sbjct: 164 KLAQIRHIYHSELEKYEQACNEFTTHVMNLLREQSRTRPVAPKEMERMVSIIHRKFSAIQ 223
Query: 380 LLTNQKTLDLIMILNSKRFLDRLVSSLEEKKHHEVKLREGLGDLSVKRMELQNALSSSWP 201
+ Q T + +MIL S RFLD + ++++ + E L + + LS+ +P
Sbjct: 224 MQLKQSTCEAVMILRS-RFLD----ARRKRRNFSKQATEVLNEY------FYSHLSNPYP 272
Query: 200 KQEAAITKTRELKKLCESTLSSV 132
+EA EL K C T+S V
Sbjct: 273 SEEA----KEELAKKCGITVSQV 291
>sptr|Q9BTW4|Q9BTW4 Similar to pre-B-cell leukemia transcription
factor 2.
Length = 349
Score = 33.9 bits (76), Expect = 1.6
Identities = 40/143 (27%), Positives = 66/143 (46%), Gaps = 15/143 (10%)
Frame = -2
Query: 515 RLGEMRNADTSSLQHQVQAVSPF------VLQQH------APDSLENMLVEIS---SAIS 381
+L ++R+ S L+ QA + F +L++ AP +E M+ I SAI
Sbjct: 164 KLAQIRHIYHSELEKYEQACNEFTTHVMNLLREQSRTRPVAPKEMERMVSIIHRKFSAIQ 223
Query: 380 LLTNQKTLDLIMILNSKRFLDRLVSSLEEKKHHEVKLREGLGDLSVKRMELQNALSSSWP 201
+ Q T + +MIL S RFLD + ++++ + E L + + LS+ +P
Sbjct: 224 MQLKQSTCEAVMILRS-RFLD----ARRKRRNFSKQATEVLNEY------FYSHLSNPYP 272
Query: 200 KQEAAITKTRELKKLCESTLSSV 132
+EA EL K C T+S V
Sbjct: 273 SEEA----KEELAKKCGITVSQV 291
>sw|Q59037|SMC_METJA Chromosome partition protein smc homolog.
Length = 1169
Score = 33.5 bits (75), Expect = 2.1
Identities = 30/144 (20%), Positives = 71/144 (49%), Gaps = 7/144 (4%)
Frame = -2
Query: 572 EYRNNILDDLLEVKSF-----LTQRLGEMR--NADTSSLQHQVQAVSPFVLQQHAPDSLE 414
E R I+++L E +S + + GE++ + + L++++ V + P
Sbjct: 781 ERREKIINELKEYESDENLKRMNEIEGELKILEKEKAKLKNEIDKGLTLVKEILIPK--- 837
Query: 413 NMLVEISSAISLLTNQKTLDLIMILNSKRFLDRLVSSLEEKKHHEVKLREGLGDLSVKRM 234
+ E++ +S L N+K + I K +++ +S LEEK+ +L + L +L+ K+
Sbjct: 838 --IEELNKKVSELINKKVILEKNISFYKESIEKNLSILEEKRKRYEELAKNLKELTEKKE 895
Query: 233 ELQNALSSSWPKQEAAITKTRELK 162
+L+ + + ++ + K R+++
Sbjct: 896 QLEKEIETLERERREILRKVRDIE 919
>sptr|O28714|O28714 Chromosome segregation protein (SMC1).
Length = 1156
Score = 33.5 bits (75), Expect = 2.1
Identities = 36/153 (23%), Positives = 66/153 (43%), Gaps = 8/153 (5%)
Frame = -2
Query: 602 AEDRSLLLEKEYRNNILDDLLEVKSFLTQRLGEMRNADTSSLQHQVQAVSPFVLQQHAPD 423
AE R +L+ +E LLE + L+ ++ E++ +A S + D
Sbjct: 657 AEKRGILVSRE--------LLEKERMLSDKIYELQREKEGLFAELNRAESLRKQYKDEVD 708
Query: 422 SLENMLVEISSAISLL-----TNQKTLDLIM--ILNSKRFLDRLVSSLEEKKHHEVKLRE 264
L M+ E+ + ISLL T ++ + I R + +SSL++ ++ E
Sbjct: 709 RLTGMISELRNRISLLDEKIRTESGRIEELREKISQKSREKENYISSLKDYNSKLAEMEE 768
Query: 263 GLGDLSVKRMELQNAL-SSSWPKQEAAITKTRE 168
+G+L + E++ L S PK + K +E
Sbjct: 769 AIGELEAEIEEIERMLRGSEVPKIVEELDKIKE 801
>sptr|O29716|O29716 Hypothetical protein AF0534.
Length = 319
Score = 33.5 bits (75), Expect = 2.1
Identities = 21/67 (31%), Positives = 35/67 (52%), Gaps = 2/67 (2%)
Frame = -2
Query: 398 ISSAISLLTNQKTLDLIMIL--NSKRFLDRLVSSLEEKKHHEVKLREGLGDLSVKRMELQ 225
+SS + L + LD+ ++L + +R+ D E KKHH +LRE L L+ + +
Sbjct: 203 LSSYLQSLEKVENLDVKLVLPGHRRRWNDLKTRVEEIKKHHSKRLREALSALNTPKTAWE 262
Query: 224 NALSSSW 204
A S +W
Sbjct: 263 VAPSITW 269
>sptr|Q9Z1J0|Q9Z1J0 Kinesin-related mitotic motor protein
(Fragment).
Length = 1014
Score = 33.1 bits (74), Expect = 2.8
Identities = 40/155 (25%), Positives = 66/155 (42%), Gaps = 6/155 (3%)
Frame = -2
Query: 614 SQMIAEDRSLLLE-KEYRNNILDDLL-----EVKSFLTQRLGEMRNADTSSLQHQVQAVS 453
S MI E++SL + K ++D+L+ +K+ + + + N + LQH +A S
Sbjct: 565 SDMILEEQSLAAQSKSVLQGLIDELVTDLFTSLKTIVAPSVVSILNIN-KQLQHIFRASS 623
Query: 452 PFVLQQHAPDSLENMLVEISSAISLLTNQKTLDLIMILNSKRFLDRLVSSLEEKKHHEVK 273
+ +E+ EI S +S+L N N + VSSL E +
Sbjct: 624 TVA------EKVEDQKREIDSFLSILCN----------NLHELRENTVSSLVESQKLCGD 667
Query: 272 LREGLGDLSVKRMELQNALSSSWPKQEAAITKTRE 168
L E L + + LSSSW ++ A+ K E
Sbjct: 668 LTEDLKTIKETHSQELCQLSSSWAERFCALEKKYE 702
>sptr|Q8KWD3|Q8KWD3 RB109.
Length = 503
Score = 32.7 bits (73), Expect = 3.7
Identities = 32/164 (19%), Positives = 67/164 (40%)
Frame = -2
Query: 617 QSQMIAEDRSLLLEKEYRNNILDDLLEVKSFLTQRLGEMRNADTSSLQHQVQAVSPFVLQ 438
++Q+ D L E+E+ D+LL+ T +L + A+ + ++ ++A +
Sbjct: 315 EAQLGQADAERLAEREHAARKYDELLQKMQAETAQLQAVMQAEKAQMEAAIEAEK---TR 371
Query: 437 QHAPDSLENMLVEISSAISLLTNQKTLDLIMILNSKRFLDRLVSSLEEKKHHEVKLREGL 258
A E +E + Q L L+ + K + L ++L ++ H +E L
Sbjct: 372 MEAAIEAEKTRMEAAIEAEKTKAQTRLRLMQVEQQKAYEVDLSTALSGQRRHAEMRQEEL 431
Query: 257 GDLSVKRMELQNALSSSWPKQEAAITKTRELKKLCESTLSSVFD 126
R E Q A+++ Q + + E+++L + D
Sbjct: 432 ------RAEWQRAVAARDQAQALVVHRDEEIERLRRDLERQIID 469
>sptrnew|AAP17974|AAP17974 Heat shock protein.
Length = 857
Score = 32.7 bits (73), Expect = 3.7
Identities = 40/202 (19%), Positives = 76/202 (37%), Gaps = 32/202 (15%)
Frame = -2
Query: 572 EYRNNILDDLLEVKSFLTQRLGEMRNADTSS----------LQHQVQAVSP--------- 450
EYR I D + F + E DT + L H VQ P
Sbjct: 318 EYRQYIEKDAALERRFQKVFVAEPSVEDTIAILRGLKERYELHHHVQITDPAIVAAATLS 377
Query: 449 --FVLQQHAPDSLENMLVEISSAISLLTNQKTLDLIMILNSKRFLDRLVSSLEEKKHHEV 276
++ + PD +++ E +S+I + + K +L DRL + + K +
Sbjct: 378 HRYIADRQLPDKAIDLIDEAASSIRMQIDSKPEEL----------DRLDRRIIQLKLEQQ 427
Query: 275 KLREGLGDLSVKRMELQN-----------ALSSSWPKQEAAITKTRELKKLCESTLSSVF 129
L + + S KR+++ N L W ++A+++ T+ +K E ++
Sbjct: 428 ALMKESDEASKKRLDMLNEELSDKERQYSELEEEWKAEKASLSGTQTIKAELEQAKIAIE 487
Query: 128 DGRPVYIIGEINTLLSSSVSQL 63
R V + ++ L + +L
Sbjct: 488 QARRVGDLARMSELQYGKIPEL 509
Database: /db/trembl-ebi/tmp/swall
Posted date: Sep 26, 2003 8:29 PM
Number of letters in database: 406,886,216
Number of sequences in database: 1,272,877
Lambda K H
0.318 0.135 0.401
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 379,232,707
Number of Sequences: 1272877
Number of extensions: 6345780
Number of successful extensions: 19083
Number of sequences better than 10.0: 30
Number of HSP's better than 10.0 without gapping: 18402
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 19031
length of database: 406,886,216
effective HSP length: 117
effective length of database: 257,959,607
effective search space used: 23474324237
frameshift window, decay const: 50, 0.1
T: 12
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)

