BLASTX 2.2.6 [Apr-09-2003]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= 2404801.2.3
(1028 letters)
Database: /db/trembl-ebi/tmp/swall
1,288,562 sequences; 411,996,502 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
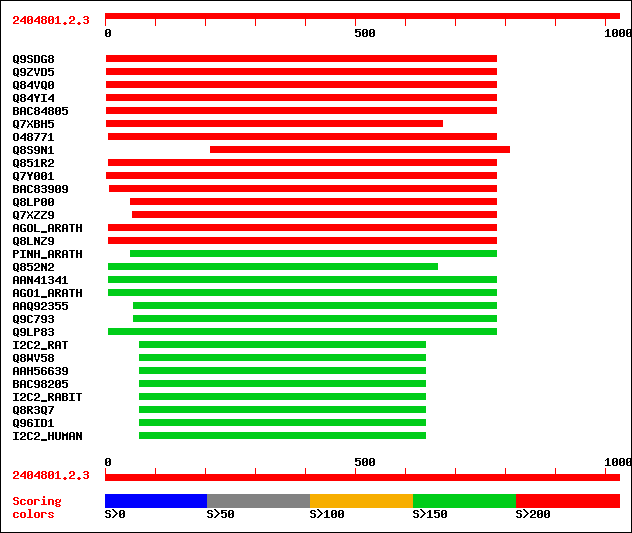
sptr|Q9SDG8|Q9SDG8 ESTs AU068544(C30430). 424 e-118
sptr|Q9ZVD5|Q9ZVD5 Argonaute (AGO1)-like protein (Putative post-... 380 e-104
sptr|Q84VQ0|Q84VQ0 PAZ (Piwi Argonaut and Zwille) family. 376 e-103
sptr|Q84YI4|Q84YI4 ARGONAUTE9 protein. 374 e-102
sptrnew|BAC84805|BAC84805 Putative ARGONAUTE9 protein. 345 8e-94
sptr|Q7XBH5|Q7XBH5 Zwille pinhead-like protein. 322 4e-87
sptr|O48771|O48771 Argonaute (AGO1)-like protein. 288 7e-77
sptr|Q8S9N1|Q8S9N1 Zwille/pinhead-like protein (Fragment). 253 4e-66
sptr|Q851R2|Q851R2 Putative argonaute protein. 213 4e-54
sptr|Q7Y001|Q7Y001 Putative leaf development and shoot apical me... 208 9e-53
sptrnew|BAC83909|BAC83909 Putative leaf development protein Argo... 207 2e-52
sptr|Q8LP00|Q8LP00 ZLL/PNH homologous protein. 206 4e-52
sptr|Q7XZZ9|Q7XZZ9 Putative leaf development and shoot apical me... 206 6e-52
sw|Q9SJK3|AGOL_ARATH Argonaute-like protein At2g27880. 203 4e-51
sptr|Q8LNZ9|Q8LNZ9 AGO1 homologous protein. 202 8e-51
sw|Q9XGW1|PINH_ARATH PINHEAD protein (ZWILLE protein). 199 5e-50
sptr|Q852N2|Q852N2 Putative argonaute protein. 198 9e-50
sptrnew|AAN41341|AAN41341 Putative leaf development protein Argo... 196 3e-49
sw|O04379|AGO1_ARATH Argonaute protein. 196 3e-49
sptrnew|AAQ92355|AAQ92355 ZIPPY. 191 2e-47
sptr|Q9C793|Q9C793 Pinhead-like protein. 191 2e-47
sptr|Q9LP83|Q9LP83 T1N15.2. 184 2e-45
sw|Q9QZ81|I2C2_RAT Eukaryotic translation initiation factor 2C 2... 178 1e-43
sptr|Q8WV58|Q8WV58 Hypothetical protein (Eukaryotic translation ... 177 2e-43
sptrnew|AAH56639|AAH56639 Hypothetical protein (Fragment). 177 2e-43
sptrnew|BAC98205|BAC98205 MKIAA1567 protein (Fragment). 177 2e-43
sw|O77503|I2C2_RABIT Eukaryotic translation initiation factor 2C... 177 2e-43
sptr|Q8R3Q7|Q8R3Q7 Similar to eukaryotic translation initiation ... 177 2e-43
sptr|Q96ID1|Q96ID1 Hypothetical protein. 177 2e-43
sw|Q9UKV8|I2C2_HUMAN Eukaryotic translation initiation factor 2C... 177 2e-43
>sptr|Q9SDG8|Q9SDG8 ESTs AU068544(C30430).
Length = 904
Score = 424 bits (1091), Expect = e-118
Identities = 206/261 (78%), Positives = 217/261 (83%), Gaps = 1/261 (0%)
Frame = +3
Query: 3 SPGHXXXXXXXXXXXXREWPLISKYRASVRTQSPKMEMIDSLFKPREA-EDDGLIRECLI 179
SPG REWPL+SKYRASVR+QSPK+EMID LFKP+ A EDDGLIRE L+
Sbjct: 644 SPGQSDIPSIAAVVSSREWPLVSKYRASVRSQSPKLEMIDGLFKPQGAQEDDGLIRELLV 703
Query: 180 DFYTSSGKRKPDQVIIFRDGVSESQFNQVLNIELQQIIEACKFLDEKWNPKFTLIIAQKN 359
DFYTS+GKRKPDQVIIFRDGVSESQF QVLNIEL QIIEACKFLDE W+PKFTLI+AQKN
Sbjct: 704 DFYTSTGKRKPDQVIIFRDGVSESQFTQVLNIELDQIIEACKFLDENWSPKFTLIVAQKN 763
Query: 360 HHTKFFIPGKPDNVPPGTVVDNKVCHPKNFDFYMCAHAGMIGTTRPTHYHILHDEIGFSP 539
HHTKFF+PG +NVPPGTVVDN VCHP+N DFYMCAHAGMIGTTRPTHYHILHDEIGFS
Sbjct: 764 HHTKFFVPGSQNNVPPGTVVDNAVCHPRNNDFYMCAHAGMIGTTRPTHYHILHDEIGFSA 823
Query: 540 DDLQELVHSLSYVYQRSTTAISVVAPICYAHLAAAQVGQFIKFDEMXXXXXXXXXXXXXX 719
DDLQELVHSLSYVYQRSTTAISVVAPICYAHLAAAQV QFIKFDEM
Sbjct: 824 DDLQELVHSLSYVYQRSTTAISVVAPICYAHLAAAQVSQFIKFDEMSETSSSHGGHTSAG 883
Query: 720 XVPVQELPRLHEKVRSSMFFC 782
PV ELPRLH KVRSSMFFC
Sbjct: 884 SAPVPELPRLHNKVRSSMFFC 904
>sptr|Q9ZVD5|Q9ZVD5 Argonaute (AGO1)-like protein (Putative
post-transcriptional gene silencing protein).
Length = 924
Score = 380 bits (977), Expect = e-104
Identities = 179/260 (68%), Positives = 204/260 (78%)
Frame = +3
Query: 3 SPGHXXXXXXXXXXXXREWPLISKYRASVRTQSPKMEMIDSLFKPREAEDDGLIRECLID 182
SPG REWPLISKYRASVRTQ K EMI+SL K EDDG+I+E L+D
Sbjct: 665 SPGQSDVPSIAAVVSSREWPLISKYRASVRTQPSKAEMIESLVKKNGTEDDGIIKELLVD 724
Query: 183 FYTSSGKRKPDQVIIFRDGVSESQFNQVLNIELQQIIEACKFLDEKWNPKFTLIIAQKNH 362
FYTSS KRKP+ +IIFRDGVSESQFNQVLNIEL QIIEACK LD WNPKF L++AQKNH
Sbjct: 725 FYTSSNKRKPEHIIIFRDGVSESQFNQVLNIELDQIIEACKLLDANWNPKFLLLVAQKNH 784
Query: 363 HTKFFIPGKPDNVPPGTVVDNKVCHPKNFDFYMCAHAGMIGTTRPTHYHILHDEIGFSPD 542
HTKFF P P+NVPPGT++DNK+CHPKN DFY+CAHAGMIGTTRPTHYH+L+DEIGFS D
Sbjct: 785 HTKFFQPTSPENVPPGTIIDNKICHPKNNDFYLCAHAGMIGTTRPTHYHVLYDEIGFSAD 844
Query: 543 DLQELVHSLSYVYQRSTTAISVVAPICYAHLAAAQVGQFIKFDEMXXXXXXXXXXXXXXX 722
+LQELVHSLSYVYQRST+AISVVAPICYAHLAAAQ+G F+KF++
Sbjct: 845 ELQELVHSLSYVYQRSTSAISVVAPICYAHLAAAQLGTFMKFEDQSETSSSHGGITAPGP 904
Query: 723 VPVQELPRLHEKVRSSMFFC 782
+ V +LPRL + V +SMFFC
Sbjct: 905 ISVAQLPRLKDNVANSMFFC 924
>sptr|Q84VQ0|Q84VQ0 PAZ (Piwi Argonaut and Zwille) family.
Length = 892
Score = 376 bits (965), Expect = e-103
Identities = 172/260 (66%), Positives = 207/260 (79%)
Frame = +3
Query: 3 SPGHXXXXXXXXXXXXREWPLISKYRASVRTQSPKMEMIDSLFKPREAEDDGLIRECLID 182
SPG R+WPLISKY+A VRTQS KMEMID+LFKP +D+G+ RE L+D
Sbjct: 633 SPGQSDIPSIAAVVSSRQWPLISKYKACVRTQSRKMEMIDNLFKPVNGKDEGMFRELLLD 692
Query: 183 FYTSSGKRKPDQVIIFRDGVSESQFNQVLNIELQQIIEACKFLDEKWNPKFTLIIAQKNH 362
FY SS RKP+ +IIFRDGVSESQFNQVLNIEL Q+++ACKFLD+ W+PKFT+I+AQKNH
Sbjct: 693 FYYSSENRKPEHIIIFRDGVSESQFNQVLNIELDQMMQACKFLDDTWHPKFTVIVAQKNH 752
Query: 363 HTKFFIPGKPDNVPPGTVVDNKVCHPKNFDFYMCAHAGMIGTTRPTHYHILHDEIGFSPD 542
HTKFF PDNVPPGT++D+++CHP+NFDFY+CAHAGMIGTTRPTHYH+L+DEIGF+ D
Sbjct: 753 HTKFFQSRGPDNVPPGTIIDSQICHPRNFDFYLCAHAGMIGTTRPTHYHVLYDEIGFATD 812
Query: 543 DLQELVHSLSYVYQRSTTAISVVAPICYAHLAAAQVGQFIKFDEMXXXXXXXXXXXXXXX 722
DLQELVHSLSYVYQRSTTAISVVAP+CYAHLAAAQ+G +K++E+
Sbjct: 813 DLQELVHSLSYVYQRSTTAISVVAPVCYAHLAAAQMGTVMKYEELSETSSSHGGITTPGA 872
Query: 723 VPVQELPRLHEKVRSSMFFC 782
VPV +P+LH V +SMFFC
Sbjct: 873 VPVPPMPQLHNNVSTSMFFC 892
>sptr|Q84YI4|Q84YI4 ARGONAUTE9 protein.
Length = 896
Score = 374 bits (960), Expect = e-102
Identities = 171/260 (65%), Positives = 207/260 (79%)
Frame = +3
Query: 3 SPGHXXXXXXXXXXXXREWPLISKYRASVRTQSPKMEMIDSLFKPREAEDDGLIRECLID 182
SPG R+WPLISKY+A VRTQS KMEMID+LFKP +D+G+ RE L+D
Sbjct: 637 SPGQSDIPSIAAVVSSRQWPLISKYKACVRTQSRKMEMIDNLFKPVNGKDEGMFRELLLD 696
Query: 183 FYTSSGKRKPDQVIIFRDGVSESQFNQVLNIELQQIIEACKFLDEKWNPKFTLIIAQKNH 362
FY SS RKP+ +IIFRDGVSESQFNQVLNIEL Q+++ACKFLD+ W+PKFT+I+AQKNH
Sbjct: 697 FYYSSENRKPEHIIIFRDGVSESQFNQVLNIELDQMMQACKFLDDTWHPKFTVIVAQKNH 756
Query: 363 HTKFFIPGKPDNVPPGTVVDNKVCHPKNFDFYMCAHAGMIGTTRPTHYHILHDEIGFSPD 542
HTKFF PDNVPPGT++D+++CHP+NFDFY+CAHAGMIGTTRPTHYH+L+DEIGF+ D
Sbjct: 757 HTKFFQSRGPDNVPPGTIIDSQICHPRNFDFYLCAHAGMIGTTRPTHYHVLYDEIGFATD 816
Query: 543 DLQELVHSLSYVYQRSTTAISVVAPICYAHLAAAQVGQFIKFDEMXXXXXXXXXXXXXXX 722
DLQELVHSLS+VYQRSTTAISVVAP+CYAHLAAAQ+G +K++E+
Sbjct: 817 DLQELVHSLSHVYQRSTTAISVVAPVCYAHLAAAQMGTVMKYEELSETSSSHGGITTPGT 876
Query: 723 VPVQELPRLHEKVRSSMFFC 782
VPV +P+LH V +SMFFC
Sbjct: 877 VPVPPMPQLHNNVSTSMFFC 896
>sptrnew|BAC84805|BAC84805 Putative ARGONAUTE9 protein.
Length = 889
Score = 345 bits (884), Expect = 8e-94
Identities = 172/261 (65%), Positives = 200/261 (76%), Gaps = 1/261 (0%)
Frame = +3
Query: 3 SPGHXXXXXXXXXXXXREWPLISKYRASVRTQSPKMEMIDSLFKPRE-AEDDGLIRECLI 179
SPG R WPLIS+YRASVRTQSPK+EMIDSLFKP + +DDG+IRE L+
Sbjct: 629 SPGRADVPSIAAVVGSRCWPLISRYRASVRTQSPKVEMIDSLFKPLDDGKDDGIIRELLL 688
Query: 180 DFYTSSGKRKPDQVIIFRDGVSESQFNQVLNIELQQIIEACKFLDEKWNPKFTLIIAQKN 359
DFY +S +RKP Q+IIFRDGVSESQF+QVLN+EL QII+A +++D+ PKFT+IIAQKN
Sbjct: 689 DFYKTSQQRKPKQIIIFRDGVSESQFSQVLNVELNQIIKAYQYMDQGPIPKFTVIIAQKN 748
Query: 360 HHTKFFIPGKPDNVPPGTVVDNKVCHPKNFDFYMCAHAGMIGTTRPTHYHILHDEIGFSP 539
HHTK F PDNVPPGTVVD+ + HP+ +DFYM AHAG IGT+RPTHYH+L DEIGF P
Sbjct: 749 HHTKLFQENTPDNVPPGTVVDSGIVHPRQYDFYMYAHAGPIGTSRPTHYHVLLDEIGFLP 808
Query: 540 DDLQELVHSLSYVYQRSTTAISVVAPICYAHLAAAQVGQFIKFDEMXXXXXXXXXXXXXX 719
DD+Q+LV SLSYVYQRSTTAISVVAPICYAHLAAAQ+GQF+KF+E
Sbjct: 809 DDVQKLVLSLSYVYQRSTTAISVVAPICYAHLAAAQMGQFMKFEEFAETSSGSGGVPSSS 868
Query: 720 XVPVQELPRLHEKVRSSMFFC 782
V ELPRLH V SSMFFC
Sbjct: 869 GAVVPELPRLHADVCSSMFFC 889
>sptr|Q7XBH5|Q7XBH5 Zwille pinhead-like protein.
Length = 361
Score = 322 bits (826), Expect = 4e-87
Identities = 158/225 (70%), Positives = 186/225 (82%), Gaps = 1/225 (0%)
Frame = +3
Query: 3 SPGHXXXXXXXXXXXXREWPLISKYRASVRTQSPKMEMIDSLFKPRE-AEDDGLIRECLI 179
SPG R WPLIS+YRASVRTQSPK+EMIDSLFKP + +DDG+IRE L+
Sbjct: 103 SPGRADVPSIAAVAGSRCWPLISRYRASVRTQSPKVEMIDSLFKPLDDGKDDGIIRELLL 162
Query: 180 DFYTSSGKRKPDQVIIFRDGVSESQFNQVLNIELQQIIEACKFLDEKWNPKFTLIIAQKN 359
DFY +S +RKP Q+IIFRDGVSESQF+QVLN+EL QII+A +++D+ PKFT+IIAQKN
Sbjct: 163 DFYKTSQQRKPKQIIIFRDGVSESQFSQVLNVELNQIIKAYQYMDQGPIPKFTVIIAQKN 222
Query: 360 HHTKFFIPGKPDNVPPGTVVDNKVCHPKNFDFYMCAHAGMIGTTRPTHYHILHDEIGFSP 539
HHTK F PDNVPPGTVVD+ + HP+ +DFYM AHAG IGT+RPTHYH+L DEIGF P
Sbjct: 223 HHTKLFQENTPDNVPPGTVVDSGIVHPRQYDFYMYAHAGPIGTSRPTHYHVLLDEIGFLP 282
Query: 540 DDLQELVHSLSYVYQRSTTAISVVAPICYAHLAAAQVGQFIKFDE 674
DD+Q+LV SLSYVYQRSTTAISVVAPICYAHLAAAQ+GQF+KF+E
Sbjct: 283 DDVQKLVLSLSYVYQRSTTAISVVAPICYAHLAAAQMGQFMKFEE 327
>sptr|O48771|O48771 Argonaute (AGO1)-like protein.
Length = 887
Score = 288 bits (738), Expect = 7e-77
Identities = 151/270 (55%), Positives = 180/270 (66%), Gaps = 11/270 (4%)
Frame = +3
Query: 6 PGHXXXXXXXXXXXXREWPLISKYRASVRTQSPKMEMIDSLFKP---REAEDDGLIRECL 176
PG + WPLIS+YRA+VRTQSP++EMIDSLF+P E D+G++ E
Sbjct: 630 PGRADVPSVAAVVGSKCWPLISRYRAAVRTQSPRLEMIDSLFQPIENTEKGDNGIMNELF 689
Query: 177 IDFYTSSGKRKPDQVIIFRDGVSESQFNQVLNIELQQIIEACKFLDEKWNPKFTLIIAQK 356
++FY +S RKP Q+IIFRDGVSESQF QVL IE+ QII+A + L E PKFT+I+AQK
Sbjct: 690 VEFYRTSRARKPKQIIIFRDGVSESQFEQVLKIEVDQIIKAYQRLGESDVPKFTVIVAQK 749
Query: 357 NHHTKFFIPGKPDNVPPGTVVDNKVCHPKNFDFYMCAHAGMIGTTRPTHYHILHDEIGFS 536
NHHTK F P+NVP GTVVD K+ HP N+DFYMCAHAG IGT+RP HYH+L DEIGFS
Sbjct: 750 NHHTKLFQAKGPENVPAGTVVDTKIVHPTNYDFYMCAHAGKIGTSRPAHYHVLLDEIGFS 809
Query: 537 PDDLQELVHSLSYVYQRSTTAIS--------VVAPICYAHLAAAQVGQFIKFDEMXXXXX 692
PDDLQ L+HSLSY S +S VAP+ YAHLAAAQV QF KF+ +
Sbjct: 810 PDDLQNLIHSLSYKLLNSIFNVSSLLCVFVLSVAPVRYAHLAAAQVAQFTKFEGISEDG- 868
Query: 693 XXXXXXXXXXVPVQELPRLHEKVRSSMFFC 782
V ELPRLHE V +MFFC
Sbjct: 869 -----------KVPELPRLHENVEGNMFFC 887
>sptr|Q8S9N1|Q8S9N1 Zwille/pinhead-like protein (Fragment).
Length = 193
Score = 253 bits (645), Expect = 4e-66
Identities = 130/200 (65%), Positives = 150/200 (75%)
Frame = +3
Query: 210 PDQVIIFRDGVSESQFNQVLNIELQQIIEACKFLDEKWNPKFTLIIAQKNHHTKFFIPGK 389
P Q+IIFRDGVSESQF+QVLN+EL QII+A +++D+ PKFT+IIAQKNHHTK F
Sbjct: 1 PKQIIIFRDGVSESQFSQVLNVELNQIIKAYQYMDQGPIPKFTVIIAQKNHHTKLFQENT 60
Query: 390 PDNVPPGTVVDNKVCHPKNFDFYMCAHAGMIGTTRPTHYHILHDEIGFSPDDLQELVHSL 569
PDNVPPGTVVD+ + HP+ +DFYM AHAG IGT+RPTHYH+L DEIGF PDD+Q+LV SL
Sbjct: 61 PDNVPPGTVVDSGIVHPRQYDFYMYAHAGPIGTSRPTHYHVLLDEIGFLPDDVQKLVLSL 120
Query: 570 SYVYQRSTTAISVVAPICYAHLAAAQVGQFIKFDEMXXXXXXXXXXXXXXXVPVQELPRL 749
SYVYQRSTTAISVVAPICYAHLAAAQ+GQF+KF+E ELPRL
Sbjct: 121 SYVYQRSTTAISVVAPICYAHLAAAQMGQFMKFEEFAELRLEVVVSFVIRS-SGPELPRL 179
Query: 750 HEKVRSSMFFC*AVVLLFWW 809
H V SSM LL WW
Sbjct: 180 HADVCSSM------SLLRWW 193
>sptr|Q851R2|Q851R2 Putative argonaute protein.
Length = 1058
Score = 213 bits (542), Expect = 4e-54
Identities = 111/274 (40%), Positives = 157/274 (57%), Gaps = 15/274 (5%)
Frame = +3
Query: 6 PGHXXXXXXXXXXXXREWPLISKYRASVRTQSPKMEMIDSLFK----PREAEDDGLIREC 173
PG +WP I+KYR V Q + E+I+ LF P + + G+IRE
Sbjct: 786 PGEDSASSIAAVVASMDWPEITKYRGLVSAQPHRQEIIEDLFSVGKDPVKVVNGGMIREL 845
Query: 174 LIDFYTSSGKRKPDQVIIFRDGVSESQFNQVLNIELQQIIEACKFLDEKWNPKFTLIIAQ 353
LI F +G+R P+++I +RDGVSE QF+ VL E+ I +AC L+E + P T ++ Q
Sbjct: 846 LIAFRKKTGRR-PERIIFYRDGVSEGQFSHVLLHEMDAIRKACASLEEGYLPPVTFVVVQ 904
Query: 354 KNHHTKFF--------IPGKPDNVPPGTVVDNKVCHPKNFDFYMCAHAGMIGTTRPTHYH 509
K HHT+ F + K N+ PGTVVD ++CHP FDFY+C+HAG+ GT+RPTHYH
Sbjct: 905 KRHHTRLFPEVHGRRDMTDKSGNILPGTVVDRQICHPTEFDFYLCSHAGIQGTSRPTHYH 964
Query: 510 ILHDEIGFSPDDLQELVHSLSYVYQRSTTAISVVAPICYAHLAAAQVGQFIK---FDEMX 680
+L+DE F+ D LQ L ++L Y Y R T A+SVV P YAHLAA + +++ D
Sbjct: 965 VLYDENHFTADALQSLTNNLCYTYARCTRAVSVVPPAYYAHLAAFRARYYVEGESSDGGS 1024
Query: 681 XXXXXXXXXXXXXXVPVQELPRLHEKVRSSMFFC 782
V V++LP++ E V+ MF+C
Sbjct: 1025 TPGSSGQAVAREGPVEVRQLPKIKENVKDVMFYC 1058
>sptr|Q7Y001|Q7Y001 Putative leaf development and shoot apical
meristem regulating protein.
Length = 1055
Score = 208 bits (530), Expect = 9e-53
Identities = 108/273 (39%), Positives = 153/273 (56%), Gaps = 13/273 (4%)
Frame = +3
Query: 3 SPGHXXXXXXXXXXXXREWPLISKYRASVRTQSPKMEMIDSLFKPREAEDD-------GL 161
SPG +WP ++KY+ V TQS + E+I +L+ E +D G+
Sbjct: 788 SPGEDASPSIAAVVASMDWPEVTKYKCLVSTQSHREEIISNLYT--EVKDPLKGIIRGGM 845
Query: 162 IRECLIDFYTSSGKRKPDQVIIFRDGVSESQFNQVLNIELQQIIEACKFLDEKWNPKFTL 341
IRE L FY +G+ KP ++I +RDG+SE QF+QVL E+ I +AC L E + P T
Sbjct: 846 IRELLRSFYQETGQ-KPSRIIFYRDGISEGQFSQVLLYEMDAIRKACASLQEGYLPPVTF 904
Query: 342 IIAQKNHHTKFFIPGKPD------NVPPGTVVDNKVCHPKNFDFYMCAHAGMIGTTRPTH 503
++ QK HHT+ F + D N+ PGTVVD +CHP FDFY+C+H+G+ GT+RPTH
Sbjct: 905 VVVQKRHHTRLFPENRRDMMDRSGNILPGTVVDTMICHPSEFDFYLCSHSGIKGTSRPTH 964
Query: 504 YHILHDEIGFSPDDLQELVHSLSYVYQRSTTAISVVAPICYAHLAAAQVGQFIKFDEMXX 683
YH+L DE GF D LQ L ++LSY Y R T A+S+V P YAHL A + +++ +
Sbjct: 965 YHVLLDENGFKADTLQTLTYNLSYTYARCTRAVSIVPPAYYAHLGAFRARYYMEDEHSDQ 1024
Query: 684 XXXXXXXXXXXXXVPVQELPRLHEKVRSSMFFC 782
+ LP + E V+ MF+C
Sbjct: 1025 GSSSSVTTRTDRS--TKPLPEIKENVKRFMFYC 1055
>sptrnew|BAC83909|BAC83909 Putative leaf development protein
Argonaute.
Length = 1052
Score = 207 bits (528), Expect = 2e-52
Identities = 111/288 (38%), Positives = 151/288 (52%), Gaps = 30/288 (10%)
Frame = +3
Query: 9 GHXXXXXXXXXXXXREWPLISKYRASVRTQSPKMEMIDSLFKPREAEDD----------- 155
G +WP I+KY+A V Q P+ E+I LF E +
Sbjct: 765 GEDSSASIAAVVASMDWPEITKYKALVSAQPPRQEIIQDLFTMTEVAQNADAPAQKAEGS 824
Query: 156 -------GLIRECLIDFYTSSGKRKPDQVIIFRDGVSESQFNQVLNIELQQIIEACKFLD 314
G+ RE L+ FY+ + KRKP ++I +RDGVS+ QF VL E+ I +A LD
Sbjct: 825 KKNFICGGMFRELLMSFYSKNAKRKPQRIIFYRDGVSDGQFLHVLLYEMDAIKKAIASLD 884
Query: 315 EKWNPKFTLIIAQKNHHTKFF--IPGKPD------NVPPGTVVDNKVCHPKNFDFYMCAH 470
+ P T ++ QK HHT+ F + G+ D NV PGTVVD +CHP FDFY+C+H
Sbjct: 885 PAYRPLVTFVVVQKRHHTRLFPEVHGRQDLTDRSGNVRPGTVVDTNICHPSEFDFYLCSH 944
Query: 471 AGMIGTTRPTHYHILHDEIGFSPDDLQELVHSLSYVYQRSTTAISVVAPICYAHLAAAQV 650
AG+ GT+RPTHYH+LHDE FS D LQ L ++L Y Y R T ++SVV P YAHLAA +
Sbjct: 945 AGIQGTSRPTHYHVLHDENRFSADQLQMLTYNLCYTYARCTRSVSVVPPAYYAHLAAFRA 1004
Query: 651 GQFIKFDEMXXXXXXXXXXXXXXX----VPVQELPRLHEKVRSSMFFC 782
+ + M V+ LP++ E V+ MF+C
Sbjct: 1005 RYYDEPPAMDGASSVGSGGNQAAAGGQPPAVRRLPQIKENVKDVMFYC 1052
>sptr|Q8LP00|Q8LP00 ZLL/PNH homologous protein.
Length = 978
Score = 206 bits (524), Expect = 4e-52
Identities = 108/257 (42%), Positives = 150/257 (58%), Gaps = 13/257 (5%)
Frame = +3
Query: 51 REWPLISKYRASVRTQSPKMEMIDSLFKP-----REAEDDGLIRECLIDFYTSSGKRKPD 215
++WP ++KY V Q+ + E+I L+K R G+IRE LI F ++G+ KP
Sbjct: 724 QDWPEVTKYAGLVCAQAHRQELIQDLYKTWHDPQRGTVTGGMIRELLISFRKATGQ-KPL 782
Query: 216 QVIIFRDGVSESQFNQVLNIELQQIIEACKFLDEKWNPKFTLIIAQKNHHTKFFIPGKPD 395
++I +RDGVSE QF QVL EL I +AC L+ + P T ++ QK HHT+ F D
Sbjct: 783 RIIFYRDGVSEGQFYQVLLYELDAIRKACASLEPIYQPPVTFVVVQKRHHTRLFANNHKD 842
Query: 396 --------NVPPGTVVDNKVCHPKNFDFYMCAHAGMIGTTRPTHYHILHDEIGFSPDDLQ 551
N+ PGTVVD+K+CHP FDFY+C+HAG+ GT+RP HYH+L DE F+ D++Q
Sbjct: 843 RSSTDKSGNILPGTVVDSKICHPSEFDFYLCSHAGIQGTSRPAHYHVLWDENNFTADEMQ 902
Query: 552 ELVHSLSYVYQRSTTAISVVAPICYAHLAAAQVGQFIKFDEMXXXXXXXXXXXXXXXVPV 731
L ++L Y Y R T ++SVV P YAHLAA + +F EM V
Sbjct: 903 TLTNNLCYTYARCTRSVSVVPPAYYAHLAAFR-ARFYMEPEMSENQTTSKSSTGTNGTSV 961
Query: 732 QELPRLHEKVRSSMFFC 782
+ LP + EKV+ MF+C
Sbjct: 962 KPLPAVKEKVKRVMFYC 978
>sptr|Q7XZZ9|Q7XZZ9 Putative leaf development and shoot apical
meristem regulating protein.
Length = 895
Score = 206 bits (523), Expect = 6e-52
Identities = 105/263 (39%), Positives = 153/263 (58%), Gaps = 20/263 (7%)
Frame = +3
Query: 54 EWPLISKYRASVRTQSPKMEMIDSLFKPREAEDD-------GLIRECLIDFYTSSGKRKP 212
+WP +SKY+ SV +QS + E+I LF E +D G+IRE + F ++G KP
Sbjct: 635 DWPEVSKYKCSVSSQSHREEIIADLFT--EVKDSQNRLVYGGMIRELIESFRKANGSYKP 692
Query: 213 DQVIIFRDGVSESQFNQVLNIELQQIIEACKFLDEKWNPKFTLIIAQKNHHTKFFIPG-- 386
++I +RDGVSE QF+QVL E+ I +AC ++E + P T ++ QK HHT+ F
Sbjct: 693 GRIIFYRDGVSEGQFSQVLLSEMDAIRKACASIEEGYLPPVTFVVVQKRHHTRLFPEDHH 752
Query: 387 ------KPDNVPPGTVVDNKVCHPKNFDFYMCAHAGMIGTTRPTHYHILHDEIGFSPDDL 548
+ N+ PGTVVD K+CHP FDFY+C+H+G+ GT+ PTHY++L DE FS D L
Sbjct: 753 ARDQMDRSRNILPGTVVDTKICHPSEFDFYLCSHSGIQGTSHPTHYYVLFDENNFSADAL 812
Query: 549 QELVHSLSYVYQRSTTAISVVAPICYAHLAAAQVGQFIKFDEM-----XXXXXXXXXXXX 713
Q L + L Y Y R T ++S+V P+ YAHLAA++ +++ +
Sbjct: 813 QTLTYHLCYTYARCTRSVSIVPPVYYAHLAASRARHYLEEGSLPDHGSSSASAAGGSRRN 872
Query: 714 XXXVPVQELPRLHEKVRSSMFFC 782
VPV+ LP + E V+ MF+C
Sbjct: 873 DRGVPVKPLPEIKENVKQFMFYC 895
>sw|Q9SJK3|AGOL_ARATH Argonaute-like protein At2g27880.
Length = 997
Score = 203 bits (516), Expect = 4e-51
Identities = 106/272 (38%), Positives = 150/272 (55%), Gaps = 13/272 (4%)
Frame = +3
Query: 6 PGHXXXXXXXXXXXXREWPLISKYRASVRTQSPKMEMIDSLFK-----PREAEDDGLIRE 170
PG +WP I+KYR V Q+ + E+I L+K R GLIRE
Sbjct: 727 PGEDSSPSIAAVVASMDWPEINKYRGLVSAQAHREEIIQDLYKLVQDPQRGLVHSGLIRE 786
Query: 171 CLIDFYTSSGKRKPDQVIIFRDGVSESQFNQVLNIELQQIIEACKFLDEKWNPKFTLIIA 350
I F ++G+ P ++I +RDGVSE QF+QVL E+ I +AC L E + P+ T +I
Sbjct: 787 HFIAFRRATGQI-PQRIIFYRDGVSEGQFSQVLLHEMTAIRKACNSLQENYVPRVTFVIV 845
Query: 351 QKNHHTKFF--------IPGKPDNVPPGTVVDNKVCHPKNFDFYMCAHAGMIGTTRPTHY 506
QK HHT+ F + K N+ PGTVVD K+CHP FDFY+ +HAG+ GT+RP HY
Sbjct: 846 QKRHHTRLFPEQHGNRDMTDKSGNIQPGTVVDTKICHPNEFDFYLNSHAGIQGTSRPAHY 905
Query: 507 HILHDEIGFSPDDLQELVHSLSYVYQRSTTAISVVAPICYAHLAAAQVGQFIKFDEMXXX 686
H+L DE GF+ D LQ L ++L Y Y R T ++S+V P YAHLAA + +++ +
Sbjct: 906 HVLLDENGFTADQLQMLTNNLCYTYARCTKSVSIVPPAYYAHLAAFRARYYMESEMSDGG 965
Query: 687 XXXXXXXXXXXXVPVQELPRLHEKVRSSMFFC 782
+ +LP + + V+ MF+C
Sbjct: 966 SSRSRSSTTGVGQVISQLPAIKDNVKEVMFYC 997
>sptr|Q8LNZ9|Q8LNZ9 AGO1 homologous protein.
Length = 909
Score = 202 bits (513), Expect = 8e-51
Identities = 109/285 (38%), Positives = 152/285 (53%), Gaps = 26/285 (9%)
Frame = +3
Query: 6 PGHXXXXXXXXXXXXREWPLISKYRASVRTQSPKMEMIDSLFK-----PREAEDDGLIRE 170
PG ++WP ++KY V Q+ + E+I+ L+K R G+IRE
Sbjct: 626 PGEDSSPSIAAVVASQDWPEVTKYAGLVSAQAHRQELIEDLYKIWQDPQRGTVSGGMIRE 685
Query: 171 CLIDFYTSSGKRKPDQVIIFRDGVSESQFNQVLNIELQQIIEACKFLDEKWNPKFTLIIA 350
LI F S+G+ KP ++I +RDGVSE QF QVL EL I +AC L+ + PK T I+
Sbjct: 686 LLISFKRSTGE-KPQRIIFYRDGVSEGQFYQVLLYELNAIRKACASLETNYQPKVTFIVV 744
Query: 351 QKNHHTKFFIPGKPD--------NVPPGTVVDNKVCHPKNFDFYMCAHAGMIGTTRPTHY 506
QK HHT+ F D N+ PGTVVD+K+CHP FDFY+C+HAG+ GT+RP HY
Sbjct: 745 QKRHHTRLFAHNHNDQNSVDRSGNILPGTVVDSKICHPTEFDFYLCSHAGIKGTSRPAHY 804
Query: 507 HILHDEIGFSPDDLQELVHSLSYVYQRSTTAISVVAPICYAHLAAAQVGQFIKFDEMXXX 686
H+L +E F+ D LQ L ++L Y Y R T ++S+V P YAHLAA + +++ D
Sbjct: 805 HVLWNENNFTADALQILTNNLCYTYARCTRSVSIVPPAYYAHLAAFRARFYMEPDTSDSS 864
Query: 687 X-------------XXXXXXXXXXXVPVQELPRLHEKVRSSMFFC 782
V+ LP L + V+ MF+C
Sbjct: 865 SVVSGPGVRGPLSGSSTSRTRAPGGAAVKPLPALKDSVKRVMFYC 909
>sw|Q9XGW1|PINH_ARATH PINHEAD protein (ZWILLE protein).
Length = 988
Score = 199 bits (506), Expect = 5e-50
Identities = 106/260 (40%), Positives = 149/260 (57%), Gaps = 16/260 (6%)
Frame = +3
Query: 51 REWPLISKYRASVRTQSPKMEMIDSLFKP-----REAEDDGLIRECLIDFYTSSGKRKPD 215
++WP ++KY V Q+ + E+I L+K R G+IR+ LI F ++G+ KP
Sbjct: 730 QDWPEVTKYAGLVCAQAHRQELIQDLYKTWQDPVRGTVSGGMIRDLLISFRKATGQ-KPL 788
Query: 216 QVIIFRDGVSESQFNQVLNIELQQIIEACKFLDEKWNPKFTLIIAQKNHHTKFFIPGKPD 395
++I +RDGVSE QF QVL EL I +AC L+ + P T I+ QK HHT+ F D
Sbjct: 789 RIIFYRDGVSEGQFYQVLLYELDAIRKACASLEPNYQPPVTFIVVQKRHHTRLFANNHRD 848
Query: 396 --------NVPPGTVVDNKVCHPKNFDFYMCAHAGMIGTTRPTHYHILHDEIGFSPDDLQ 551
N+ PGTVVD K+CHP FDFY+C+HAG+ GT+RP HYH+L DE F+ D +Q
Sbjct: 849 KNSTDRSGNILPGTVVDTKICHPTEFDFYLCSHAGIQGTSRPAHYHVLWDENNFTADGIQ 908
Query: 552 ELVHSLSYVYQRSTTAISVVAPICYAHLAAAQVGQFIKFDEMXXXXX---XXXXXXXXXX 722
L ++L Y Y R T ++S+V P YAHLAA + +++ + M
Sbjct: 909 SLTNNLCYTYARCTRSVSIVPPAYYAHLAAFRARFYLEPEIMQDNGSPGKKNTKTTTVGD 968
Query: 723 VPVQELPRLHEKVRSSMFFC 782
V V+ LP L E V+ MF+C
Sbjct: 969 VGVKPLPALKENVKRVMFYC 988
>sptr|Q852N2|Q852N2 Putative argonaute protein.
Length = 1192
Score = 198 bits (504), Expect = 9e-50
Identities = 101/232 (43%), Positives = 142/232 (61%), Gaps = 12/232 (5%)
Frame = +3
Query: 6 PGHXXXXXXXXXXXXREWPLISKYRASVRTQSPKMEMIDSLFK----PREAEDDGLIREC 173
PG +WP I+KYR V QS + E+I+ LF P + + G+IRE
Sbjct: 540 PGEDSASSIAAVVASMDWPEITKYRGLVSAQSHRQEIIEDLFSVGKDPVKVVNGGMIREF 599
Query: 174 LIDFYTSSGKRKPDQVIIFRDGVSESQFNQVLNIELQQIIEACKFLDEKWNPKFTLIIAQ 353
LI F +G+R P+++I +RDGVSE QF++VL E+ I +AC L+E + P T ++ Q
Sbjct: 600 LIAFRKKTGRR-PERIIFYRDGVSEGQFSRVLLHEMDAIRKACASLEEGYLPPVTFVVVQ 658
Query: 354 KNHHTKFF--------IPGKPDNVPPGTVVDNKVCHPKNFDFYMCAHAGMIGTTRPTHYH 509
K HHT+ F + K N+ PGTV D ++CHP F FY+C+HAG+ GT+RPTHYH
Sbjct: 659 KRHHTRLFPEVHGRRDMTDKSGNILPGTVKDRQICHPTEFYFYLCSHAGIQGTSRPTHYH 718
Query: 510 ILHDEIGFSPDDLQELVHSLSYVYQRSTTAISVVAPICYAHLAAAQVGQFIK 665
+L+DE F+ D+LQ L ++L Y+Y R T A+SVV P Y+HLAA+ IK
Sbjct: 719 VLYDENHFTADELQTLTNNLCYIYARCTHAVSVVPPAYYSHLAASHAHCCIK 770
>sptrnew|AAN41341|AAN41341 Putative leaf development protein
Argonaute.
Length = 1048
Score = 196 bits (499), Expect = 3e-49
Identities = 108/284 (38%), Positives = 150/284 (52%), Gaps = 25/284 (8%)
Frame = +3
Query: 6 PGHXXXXXXXXXXXXREWPLISKYRASVRTQSPKMEMIDSLFKP-----REAEDDGLIRE 170
PG ++WP I+KY V Q+ + E+I LFK + G+I+E
Sbjct: 766 PGEDSSPSIAAVVASQDWPEITKYAGLVCAQAHRQELIQDLFKEWKDPQKGVVTGGMIKE 825
Query: 171 CLIDFYTSSGKRKPDQVIIFRDGVSESQFNQVLNIELQQIIEACKFLDEKWNPKFTLIIA 350
LI F S+G KP ++I +RDGVSE QF QVL EL I +AC L+ + P T ++
Sbjct: 826 LLIAFRRSTG-HKPLRIIFYRDGVSEGQFYQVLLYELDAIRKACASLEAGYQPPVTFVVV 884
Query: 351 QKNHHTKFFIPGKPD--------NVPPGTVVDNKVCHPKNFDFYMCAHAGMIGTTRPTHY 506
QK HHT+ F D N+ PGTVVD+K+CHP FDFY+C+HAG+ GT+RP HY
Sbjct: 885 QKRHHTRLFAQNHNDRHSVDRSGNILPGTVVDSKICHPTEFDFYLCSHAGIQGTSRPAHY 944
Query: 507 HILHDEIGFSPDDLQELVHSLSYVYQRSTTAISVVAPICYAHLAAAQVGQFIKFDEMXXX 686
H+L DE F+ D LQ L ++L Y Y R T ++S+V P YAHLAA + +++ +
Sbjct: 945 HVLWDENNFTADGLQSLTNNLCYTYARCTRSVSIVPPAYYAHLAAFRARFYMEPETSDSG 1004
Query: 687 XXXXXXXXXXXXV------------PVQELPRLHEKVRSSMFFC 782
+ V+ LP L E V+ MF+C
Sbjct: 1005 SMASGSMARGGGMAGRSTRGPNVNAAVRPLPALKENVKRVMFYC 1048
>sw|O04379|AGO1_ARATH Argonaute protein.
Length = 1048
Score = 196 bits (499), Expect = 3e-49
Identities = 108/284 (38%), Positives = 150/284 (52%), Gaps = 25/284 (8%)
Frame = +3
Query: 6 PGHXXXXXXXXXXXXREWPLISKYRASVRTQSPKMEMIDSLFKP-----REAEDDGLIRE 170
PG ++WP I+KY V Q+ + E+I LFK + G+I+E
Sbjct: 766 PGEDSSPSIAAVVASQDWPEITKYAGLVCAQAHRQELIQDLFKEWKDPQKGVVTGGMIKE 825
Query: 171 CLIDFYTSSGKRKPDQVIIFRDGVSESQFNQVLNIELQQIIEACKFLDEKWNPKFTLIIA 350
LI F S+G KP ++I +RDGVSE QF QVL EL I +AC L+ + P T ++
Sbjct: 826 LLIAFRRSTG-HKPLRIIFYRDGVSEGQFYQVLLYELDAIRKACASLEAGYQPPVTFVVV 884
Query: 351 QKNHHTKFFIPGKPD--------NVPPGTVVDNKVCHPKNFDFYMCAHAGMIGTTRPTHY 506
QK HHT+ F D N+ PGTVVD+K+CHP FDFY+C+HAG+ GT+RP HY
Sbjct: 885 QKRHHTRLFAQNHNDRHSVDRSGNILPGTVVDSKICHPTEFDFYLCSHAGIQGTSRPAHY 944
Query: 507 HILHDEIGFSPDDLQELVHSLSYVYQRSTTAISVVAPICYAHLAAAQVGQFIKFDEMXXX 686
H+L DE F+ D LQ L ++L Y Y R T ++S+V P YAHLAA + +++ +
Sbjct: 945 HVLWDENNFTADGLQSLTNNLCYTYARCTRSVSIVPPAYYAHLAAFRARFYMEPETSDSG 1004
Query: 687 XXXXXXXXXXXXV------------PVQELPRLHEKVRSSMFFC 782
+ V+ LP L E V+ MF+C
Sbjct: 1005 SMASGSMARGGGMAGRSTRGPNVNAAVRPLPALKENVKRVMFYC 1048
>sptrnew|AAQ92355|AAQ92355 ZIPPY.
Length = 990
Score = 191 bits (484), Expect = 2e-47
Identities = 105/245 (42%), Positives = 147/245 (60%), Gaps = 3/245 (1%)
Frame = +3
Query: 57 WPLISKYRASVRTQSPKMEMIDSLFKPREAEDDGLIRECLIDFYTSSGKRKPDQVIIFRD 236
WP ++Y + +R+Q+ + E+I L D +++E L DFY + K+ P+++I FRD
Sbjct: 757 WPEANRYVSRMRSQTHRQEIIQDL--------DLMVKELLDDFYKAV-KKLPNRIIFFRD 807
Query: 237 GVSESQFNQVLNIELQQIIEAC-KFLDEKWNPKFTLIIAQKNHHTKFF-IPGKPDNVPPG 410
GVSE+QF +VL ELQ I AC KF D +NP T + QK HHT+ F +N+PPG
Sbjct: 808 GVSETQFKKVLQEELQSIKTACSKFQD--YNPSITFAVVQKRHHTRLFRCDPDHENIPPG 865
Query: 411 TVVDNKVCHPKNFDFYMCAHAGMIGTTRPTHYHILHDEIGFSPDDLQELVHSLSYVYQRS 590
TVVD + HPK FDFY+C+H G+ GT+RPTHYHIL DE F+ D+LQ LV++L Y + R
Sbjct: 866 TVVDTVITHPKEFDFYLCSHLGVKGTSRPTHYHILWDENEFTSDELQRLVYNLCYTFVRC 925
Query: 591 TTAISVVAPICYAHLAAAQVGQFIKFDEMXXXXXXXXXXXXXXXVP-VQELPRLHEKVRS 767
T IS+V P YAHLAA + +I+ P LP+L + V++
Sbjct: 926 TKPISIVPPAYYAHLAAYRGRLYIERSSESNGGSMNPSSVSRVGPPKTIPLPKLSDNVKN 985
Query: 768 SMFFC 782
MF+C
Sbjct: 986 LMFYC 990
>sptr|Q9C793|Q9C793 Pinhead-like protein.
Length = 990
Score = 191 bits (484), Expect = 2e-47
Identities = 105/245 (42%), Positives = 147/245 (60%), Gaps = 3/245 (1%)
Frame = +3
Query: 57 WPLISKYRASVRTQSPKMEMIDSLFKPREAEDDGLIRECLIDFYTSSGKRKPDQVIIFRD 236
WP ++Y + +R+Q+ + E+I L D +++E L DFY + K+ P+++I FRD
Sbjct: 757 WPEANRYVSRMRSQTHRQEIIQDL--------DLMVKELLDDFYKAV-KKLPNRIIFFRD 807
Query: 237 GVSESQFNQVLNIELQQIIEAC-KFLDEKWNPKFTLIIAQKNHHTKFF-IPGKPDNVPPG 410
GVSE+QF +VL ELQ I AC KF D +NP T + QK HHT+ F +N+PPG
Sbjct: 808 GVSETQFKKVLQEELQSIKTACSKFQD--YNPSITFAVVQKRHHTRLFRCDPDHENIPPG 865
Query: 411 TVVDNKVCHPKNFDFYMCAHAGMIGTTRPTHYHILHDEIGFSPDDLQELVHSLSYVYQRS 590
TVVD + HPK FDFY+C+H G+ GT+RPTHYHIL DE F+ D+LQ LV++L Y + R
Sbjct: 866 TVVDTVITHPKEFDFYLCSHLGVKGTSRPTHYHILWDENEFTSDELQRLVYNLCYTFVRC 925
Query: 591 TTAISVVAPICYAHLAAAQVGQFIKFDEMXXXXXXXXXXXXXXXVP-VQELPRLHEKVRS 767
T IS+V P YAHLAA + +I+ P LP+L + V++
Sbjct: 926 TKPISIVPPAYYAHLAAYRGRLYIERSSESNGGSMNPSSVSRVGPPKTIPLPKLSDNVKN 985
Query: 768 SMFFC 782
MF+C
Sbjct: 986 LMFYC 990
>sptr|Q9LP83|Q9LP83 T1N15.2.
Length = 1123
Score = 184 bits (466), Expect = 2e-45
Identities = 107/299 (35%), Positives = 149/299 (49%), Gaps = 40/299 (13%)
Frame = +3
Query: 6 PGHXXXXXXXXXXXXREWPLISKYRASVRTQSPKMEMIDSLFKPREAEDDGLIR------ 167
PG ++WP I+KY V Q+ + E+I LFK + G++
Sbjct: 826 PGEDSSPSIAAVVASQDWPEITKYAGLVCAQAHRQELIQDLFKEWKDPQKGVVTGGMIKY 885
Query: 168 -----------ECLIDFYTSSGKRKPDQVIIFRDGVSESQFNQVLNIELQQIIEACKFLD 314
E LI F S+G KP ++I +RDGVSE QF QVL EL I +AC L+
Sbjct: 886 VWMLFNIFVIGELLIAFRRSTG-HKPLRIIFYRDGVSEGQFYQVLLYELDAIRKACASLE 944
Query: 315 EKWNPKFTLIIAQKNHHTKFFIPGKPD--------NVPPG---TVVDNKVCHPKNFDFYM 461
+ P T ++ QK HHT+ F D N+ PG TVVD+K+CHP FDFY+
Sbjct: 945 AGYQPPVTFVVVQKRHHTRLFAQNHNDRHSVDRSGNILPGETSTVVDSKICHPTEFDFYL 1004
Query: 462 CAHAGMIGTTRPTHYHILHDEIGFSPDDLQELVHSLSYVYQRSTTAISVVAPICYAHLAA 641
C+HAG+ GT+RP HYH+L DE F+ D LQ L ++L Y Y R T ++S+V P YAHLAA
Sbjct: 1005 CSHAGIQGTSRPAHYHVLWDENNFTADGLQSLTNNLCYTYARCTRSVSIVPPAYYAHLAA 1064
Query: 642 AQVGQFIKFDEMXXXXXXXXXXXXXXXV------------PVQELPRLHEKVRSSMFFC 782
+ +++ + + V+ LP L E V+ MF+C
Sbjct: 1065 FRARFYMEPETSDSGSMASGSMARGGGMAGRSTRGPNVNAAVRPLPALKENVKRVMFYC 1123
>sw|Q9QZ81|I2C2_RAT Eukaryotic translation initiation factor 2C 2
(eIF2C 2) (eIF-2C 2) (Golgi ER protein 95 kDa) (GERp95).
Length = 863
Score = 178 bits (451), Expect = 1e-43
Identities = 91/197 (46%), Positives = 125/197 (63%), Gaps = 6/197 (3%)
Frame = +3
Query: 69 SKYRASVRTQSPKMEMIDSLFKPREAEDDGLIRECLIDFYTSSGKRKPDQVIIFRDGVSE 248
++Y A+VR Q + E+I L ++RE LI FY S+ + KP ++I +RDGVSE
Sbjct: 627 NRYCATVRVQQHRQEIIQDLA--------AMVRELLIQFYKST-RFKPTRIIFYRDGVSE 677
Query: 249 SQFNQVLNIELQQIIEACKFLDEKWNPKFTLIIAQKNHHTKFFIP------GKPDNVPPG 410
QF QVL+ EL I EAC L++++ P T I+ QK HHT+ F GK N+P G
Sbjct: 678 GQFQQVLHHELLAIREACIKLEKEYQPGITFIVVQKRHHTRLFCTDKNERVGKSGNIPAG 737
Query: 411 TVVDNKVCHPKNFDFYMCAHAGMIGTTRPTHYHILHDEIGFSPDDLQELVHSLSYVYQRS 590
T VD K+ HP FDFY+C+HAG+ GT+RP+HYH+L D+ FS D+LQ L + L + Y R
Sbjct: 738 TTVDTKITHPTEFDFYLCSHAGIQGTSRPSHYHVLWDDNRFSSDELQILTYQLCHTYVRC 797
Query: 591 TTAISVVAPICYAHLAA 641
T ++S+ AP YAHL A
Sbjct: 798 TRSVSIPAPAYYAHLVA 814
>sptr|Q8WV58|Q8WV58 Hypothetical protein (Eukaryotic translation
initiation factor 2C, 2).
Length = 585
Score = 177 bits (449), Expect = 2e-43
Identities = 91/197 (46%), Positives = 124/197 (62%), Gaps = 6/197 (3%)
Frame = +3
Query: 69 SKYRASVRTQSPKMEMIDSLFKPREAEDDGLIRECLIDFYTSSGKRKPDQVIIFRDGVSE 248
++Y A+VR Q + E+I L ++RE LI FY S+ + KP ++I +RDGVSE
Sbjct: 349 NRYCATVRVQQHRQEIIQDLA--------AMVRELLIQFYKST-RFKPTRIIFYRDGVSE 399
Query: 249 SQFNQVLNIELQQIIEACKFLDEKWNPKFTLIIAQKNHHTKFFIP------GKPDNVPPG 410
QF QVL+ EL I EAC L++ + P T I+ QK HHT+ F GK N+P G
Sbjct: 400 GQFQQVLHHELLAIREACIKLEKDYQPGITFIVVQKRHHTRLFCTDKNERVGKSGNIPAG 459
Query: 411 TVVDNKVCHPKNFDFYMCAHAGMIGTTRPTHYHILHDEIGFSPDDLQELVHSLSYVYQRS 590
T VD K+ HP FDFY+C+HAG+ GT+RP+HYH+L D+ FS D+LQ L + L + Y R
Sbjct: 460 TTVDTKITHPTEFDFYLCSHAGIQGTSRPSHYHVLWDDNRFSSDELQILTYQLCHTYVRC 519
Query: 591 TTAISVVAPICYAHLAA 641
T ++S+ AP YAHL A
Sbjct: 520 TRSVSIPAPAYYAHLVA 536
>sptrnew|AAH56639|AAH56639 Hypothetical protein (Fragment).
Length = 437
Score = 177 bits (449), Expect = 2e-43
Identities = 91/197 (46%), Positives = 124/197 (62%), Gaps = 6/197 (3%)
Frame = +3
Query: 69 SKYRASVRTQSPKMEMIDSLFKPREAEDDGLIRECLIDFYTSSGKRKPDQVIIFRDGVSE 248
++Y A+VR Q + E+I L ++RE LI FY S+ + KP ++I +RDGVSE
Sbjct: 201 NRYCATVRVQQHRQEIIQDLA--------AMVRELLIQFYKST-RFKPTRIIFYRDGVSE 251
Query: 249 SQFNQVLNIELQQIIEACKFLDEKWNPKFTLIIAQKNHHTKFFIP------GKPDNVPPG 410
QF QVL+ EL I EAC L++ + P T I+ QK HHT+ F GK N+P G
Sbjct: 252 GQFQQVLHHELLAIREACIKLEKDYQPGITFIVVQKRHHTRLFCTDKNERVGKSGNIPAG 311
Query: 411 TVVDNKVCHPKNFDFYMCAHAGMIGTTRPTHYHILHDEIGFSPDDLQELVHSLSYVYQRS 590
T VD K+ HP FDFY+C+HAG+ GT+RP+HYH+L D+ FS D+LQ L + L + Y R
Sbjct: 312 TTVDTKITHPTEFDFYLCSHAGIQGTSRPSHYHVLWDDNRFSSDELQILTYQLCHTYVRC 371
Query: 591 TTAISVVAPICYAHLAA 641
T ++S+ AP YAHL A
Sbjct: 372 TRSVSIPAPAYYAHLVA 388
>sptrnew|BAC98205|BAC98205 MKIAA1567 protein (Fragment).
Length = 703
Score = 177 bits (449), Expect = 2e-43
Identities = 91/197 (46%), Positives = 124/197 (62%), Gaps = 6/197 (3%)
Frame = +3
Query: 69 SKYRASVRTQSPKMEMIDSLFKPREAEDDGLIRECLIDFYTSSGKRKPDQVIIFRDGVSE 248
++Y A+VR Q + E+I L ++RE LI FY S+ + KP ++I +RDGVSE
Sbjct: 467 NRYCATVRVQQHRQEIIQDLA--------AMVRELLIQFYKST-RFKPTRIIFYRDGVSE 517
Query: 249 SQFNQVLNIELQQIIEACKFLDEKWNPKFTLIIAQKNHHTKFFIP------GKPDNVPPG 410
QF QVL+ EL I EAC L++ + P T I+ QK HHT+ F GK N+P G
Sbjct: 518 GQFQQVLHHELLAIREACIKLEKDYQPGITFIVVQKRHHTRLFCTDKNERVGKSGNIPAG 577
Query: 411 TVVDNKVCHPKNFDFYMCAHAGMIGTTRPTHYHILHDEIGFSPDDLQELVHSLSYVYQRS 590
T VD K+ HP FDFY+C+HAG+ GT+RP+HYH+L D+ FS D+LQ L + L + Y R
Sbjct: 578 TTVDTKITHPTEFDFYLCSHAGIQGTSRPSHYHVLWDDNRFSSDELQILTYQLCHTYVRC 637
Query: 591 TTAISVVAPICYAHLAA 641
T ++S+ AP YAHL A
Sbjct: 638 TRSVSIPAPAYYAHLVA 654
>sw|O77503|I2C2_RABIT Eukaryotic translation initiation factor 2C 2
(eIF2C 2) (eIF-2C 2).
Length = 813
Score = 177 bits (449), Expect = 2e-43
Identities = 91/197 (46%), Positives = 124/197 (62%), Gaps = 6/197 (3%)
Frame = +3
Query: 69 SKYRASVRTQSPKMEMIDSLFKPREAEDDGLIRECLIDFYTSSGKRKPDQVIIFRDGVSE 248
++Y A+VR Q + E+I L ++RE LI FY S+ + KP ++I +RDGVSE
Sbjct: 577 NRYCATVRVQQHRQEIIQDLA--------AMVRELLIQFYKST-RFKPTRIIFYRDGVSE 627
Query: 249 SQFNQVLNIELQQIIEACKFLDEKWNPKFTLIIAQKNHHTKFFIP------GKPDNVPPG 410
QF QVL+ EL I EAC L++ + P T I+ QK HHT+ F GK N+P G
Sbjct: 628 GQFQQVLHHELLAIREACIKLEKDYQPGITFIVVQKRHHTRLFCTDKNERVGKSGNIPAG 687
Query: 411 TVVDNKVCHPKNFDFYMCAHAGMIGTTRPTHYHILHDEIGFSPDDLQELVHSLSYVYQRS 590
T VD K+ HP FDFY+C+HAG+ GT+RP+HYH+L D+ FS D+LQ L + L + Y R
Sbjct: 688 TTVDTKITHPTEFDFYLCSHAGIQGTSRPSHYHVLWDDNRFSSDELQILTYQLCHTYVRC 747
Query: 591 TTAISVVAPICYAHLAA 641
T ++S+ AP YAHL A
Sbjct: 748 TRSVSIPAPAYYAHLVA 764
>sptr|Q8R3Q7|Q8R3Q7 Similar to eukaryotic translation initiation
factor 2C, 2 (Fragment).
Length = 530
Score = 177 bits (449), Expect = 2e-43
Identities = 91/197 (46%), Positives = 124/197 (62%), Gaps = 6/197 (3%)
Frame = +3
Query: 69 SKYRASVRTQSPKMEMIDSLFKPREAEDDGLIRECLIDFYTSSGKRKPDQVIIFRDGVSE 248
++Y A+VR Q + E+I L ++RE LI FY S+ + KP ++I +RDGVSE
Sbjct: 294 NRYCATVRVQQHRQEIIQDLA--------AMVRELLIQFYKST-RFKPTRIIFYRDGVSE 344
Query: 249 SQFNQVLNIELQQIIEACKFLDEKWNPKFTLIIAQKNHHTKFFIP------GKPDNVPPG 410
QF QVL+ EL I EAC L++ + P T I+ QK HHT+ F GK N+P G
Sbjct: 345 GQFQQVLHHELLAIREACIKLEKDYQPGITFIVVQKRHHTRLFCTDKNERVGKSGNIPAG 404
Query: 411 TVVDNKVCHPKNFDFYMCAHAGMIGTTRPTHYHILHDEIGFSPDDLQELVHSLSYVYQRS 590
T VD K+ HP FDFY+C+HAG+ GT+RP+HYH+L D+ FS D+LQ L + L + Y R
Sbjct: 405 TTVDTKITHPTEFDFYLCSHAGIQGTSRPSHYHVLWDDNRFSSDELQILTYQLCHTYVRC 464
Query: 591 TTAISVVAPICYAHLAA 641
T ++S+ AP YAHL A
Sbjct: 465 TRSVSIPAPAYYAHLVA 481
>sptr|Q96ID1|Q96ID1 Hypothetical protein.
Length = 377
Score = 177 bits (449), Expect = 2e-43
Identities = 91/197 (46%), Positives = 124/197 (62%), Gaps = 6/197 (3%)
Frame = +3
Query: 69 SKYRASVRTQSPKMEMIDSLFKPREAEDDGLIRECLIDFYTSSGKRKPDQVIIFRDGVSE 248
++Y A+VR Q + E+I L ++RE LI FY S+ + KP ++I +RDGVSE
Sbjct: 141 NRYCATVRVQQHRQEIIQDLA--------AMVRELLIQFYKST-RFKPTRIIFYRDGVSE 191
Query: 249 SQFNQVLNIELQQIIEACKFLDEKWNPKFTLIIAQKNHHTKFFIP------GKPDNVPPG 410
QF QVL+ EL I EAC L++ + P T I+ QK HHT+ F GK N+P G
Sbjct: 192 GQFQQVLHHELLAIREACIKLEKDYQPGITFIVVQKRHHTRLFCTDKNERVGKSGNIPAG 251
Query: 411 TVVDNKVCHPKNFDFYMCAHAGMIGTTRPTHYHILHDEIGFSPDDLQELVHSLSYVYQRS 590
T VD K+ HP FDFY+C+HAG+ GT+RP+HYH+L D+ FS D+LQ L + L + Y R
Sbjct: 252 TTVDTKITHPTEFDFYLCSHAGIQGTSRPSHYHVLWDDNRFSSDELQILTYQLCHTYVRC 311
Query: 591 TTAISVVAPICYAHLAA 641
T ++S+ AP YAHL A
Sbjct: 312 TRSVSIPAPAYYAHLVA 328
>sw|Q9UKV8|I2C2_HUMAN Eukaryotic translation initiation factor 2C 2
(eIF2C 2) (eIF-2C 2) (Fragment).
Length = 377
Score = 177 bits (449), Expect = 2e-43
Identities = 91/197 (46%), Positives = 124/197 (62%), Gaps = 6/197 (3%)
Frame = +3
Query: 69 SKYRASVRTQSPKMEMIDSLFKPREAEDDGLIRECLIDFYTSSGKRKPDQVIIFRDGVSE 248
++Y A+VR Q + E+I L ++RE LI FY S+ + KP ++I +RDGVSE
Sbjct: 141 NRYCATVRVQQHRQEIIQDLA--------AMVRELLIQFYKST-RFKPTRIIFYRDGVSE 191
Query: 249 SQFNQVLNIELQQIIEACKFLDEKWNPKFTLIIAQKNHHTKFFIP------GKPDNVPPG 410
QF QVL+ EL I EAC L++ + P T I+ QK HHT+ F GK N+P G
Sbjct: 192 GQFQQVLHHELLAIREACIKLEKDYQPGITFIVVQKRHHTRLFCTDKNERVGKSGNIPAG 251
Query: 411 TVVDNKVCHPKNFDFYMCAHAGMIGTTRPTHYHILHDEIGFSPDDLQELVHSLSYVYQRS 590
T VD K+ HP FDFY+C+HAG+ GT+RP+HYH+L D+ FS D+LQ L + L + Y R
Sbjct: 252 TTVDTKITHPTEFDFYLCSHAGIQGTSRPSHYHVLWDDNRFSSDELQILTYQLCHTYVRC 311
Query: 591 TTAISVVAPICYAHLAA 641
T ++S+ AP YAHL A
Sbjct: 312 TRSVSIPAPAYYAHLVA 328
Database: /db/trembl-ebi/tmp/swall
Posted date: Oct 24, 2003 8:22 PM
Number of letters in database: 411,996,502
Number of sequences in database: 1,288,562
Lambda K H
0.318 0.135 0.401
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 787,563,927
Number of Sequences: 1288562
Number of extensions: 16971118
Number of successful extensions: 42610
Number of sequences better than 10.0: 30
Number of HSP's better than 10.0 without gapping: 40898
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 42435
length of database: 411,996,502
effective HSP length: 123
effective length of database: 253,503,376
effective search space used: 55517239344
frameshift window, decay const: 50, 0.1
T: 12
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)

