BLASTX 2.2.6 [Apr-09-2003]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= 2404801.2.1
(867 letters)
Database: /db/trembl-ebi/tmp/swall
1,288,562 sequences; 411,996,502 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
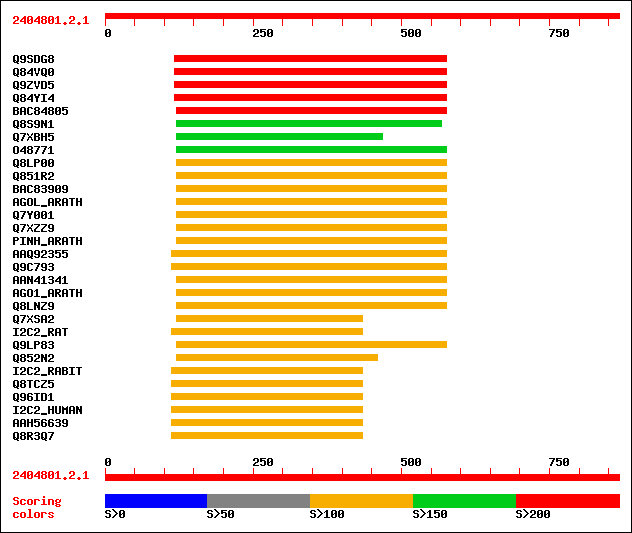
sptr|Q9SDG8|Q9SDG8 ESTs AU068544(C30430). 259 3e-68
sptr|Q84VQ0|Q84VQ0 PAZ (Piwi Argonaut and Zwille) family. 231 8e-60
sptr|Q9ZVD5|Q9ZVD5 Argonaute (AGO1)-like protein (Putative post-... 229 3e-59
sptr|Q84YI4|Q84YI4 ARGONAUTE9 protein. 229 3e-59
sptrnew|BAC84805|BAC84805 Putative ARGONAUTE9 protein. 214 1e-54
sptr|Q8S9N1|Q8S9N1 Zwille/pinhead-like protein (Fragment). 197 1e-49
sptr|Q7XBH5|Q7XBH5 Zwille pinhead-like protein. 191 9e-48
sptr|O48771|O48771 Argonaute (AGO1)-like protein. 182 5e-45
sptr|Q8LP00|Q8LP00 ZLL/PNH homologous protein. 140 2e-32
sptr|Q851R2|Q851R2 Putative argonaute protein. 140 3e-32
sptrnew|BAC83909|BAC83909 Putative leaf development protein Argo... 137 2e-31
sw|Q9SJK3|AGOL_ARATH Argonaute-like protein At2g27880. 136 3e-31
sptr|Q7Y001|Q7Y001 Putative leaf development and shoot apical me... 136 4e-31
sptr|Q7XZZ9|Q7XZZ9 Putative leaf development and shoot apical me... 134 1e-30
sw|Q9XGW1|PINH_ARATH PINHEAD protein (ZWILLE protein). 134 2e-30
sptrnew|AAQ92355|AAQ92355 ZIPPY. 133 4e-30
sptr|Q9C793|Q9C793 Pinhead-like protein. 133 4e-30
sptrnew|AAN41341|AAN41341 Putative leaf development protein Argo... 129 5e-29
sw|O04379|AGO1_ARATH Argonaute protein. 129 5e-29
sptr|Q8LNZ9|Q8LNZ9 AGO1 homologous protein. 128 9e-29
sptr|Q7XSA2|Q7XSA2 OSJNBa0005N02.3 protein. 124 1e-27
sw|Q9QZ81|I2C2_RAT Eukaryotic translation initiation factor 2C 2... 124 1e-27
sptr|Q9LP83|Q9LP83 T1N15.2. 124 2e-27
sptr|Q852N2|Q852N2 Putative argonaute protein. 123 3e-27
sw|O77503|I2C2_RABIT Eukaryotic translation initiation factor 2C... 123 3e-27
sptr|Q8TCZ5|Q8TCZ5 Eukaryotic initiation factor 2C2. 123 3e-27
sptr|Q96ID1|Q96ID1 Hypothetical protein. 123 3e-27
sw|Q9UKV8|I2C2_HUMAN Eukaryotic translation initiation factor 2C... 123 3e-27
sptrnew|AAH56639|AAH56639 Hypothetical protein (Fragment). 123 3e-27
sptr|Q8R3Q7|Q8R3Q7 Similar to eukaryotic translation initiation ... 123 3e-27
>sptr|Q9SDG8|Q9SDG8 ESTs AU068544(C30430).
Length = 904
Score = 259 bits (662), Expect = 3e-68
Identities = 122/153 (79%), Positives = 128/153 (83%)
Frame = +1
Query: 118 NPKFTLIIAQKNHHTKFFIPGKPDNVPAGTVVDNKVCHPRNFDFYMCSHAGMIGTTRPTH 297
+PKFTLI+AQKNHHTKFF+PG +NVP GTVVDN VCHPRN DFYMC+HAGMIGTTRPTH
Sbjct: 752 SPKFTLIVAQKNHHTKFFVPGSQNNVPPGTVVDNAVCHPRNNDFYMCAHAGMIGTTRPTH 811
Query: 298 YHILHDEIGFNPDDLQELVHSLSYVYQRSTTAISVVAPICYAHLAAAQVGQFIKFDEMXX 477
YHILHDEIGF+ DDLQELVHSLSYVYQRSTTAISVVAPICYAHLAAAQV QFIKFDEM
Sbjct: 812 YHILHDEIGFSADDLQELVHSLSYVYQRSTTAISVVAPICYAHLAAAQVSQFIKFDEMSE 871
Query: 478 XXXXXXXXXXXXXVPVQELPRLHEKVRSSMFFC 576
PV ELPRLH KVRSSMFFC
Sbjct: 872 TSSSHGGHTSAGSAPVPELPRLHNKVRSSMFFC 904
>sptr|Q84VQ0|Q84VQ0 PAZ (Piwi Argonaut and Zwille) family.
Length = 892
Score = 231 bits (590), Expect = 8e-60
Identities = 103/153 (67%), Positives = 124/153 (81%)
Frame = +1
Query: 118 NPKFTLIIAQKNHHTKFFIPGKPDNVPAGTVVDNKVCHPRNFDFYMCSHAGMIGTTRPTH 297
+PKFT+I+AQKNHHTKFF PDNVP GT++D+++CHPRNFDFY+C+HAGMIGTTRPTH
Sbjct: 740 HPKFTVIVAQKNHHTKFFQSRGPDNVPPGTIIDSQICHPRNFDFYLCAHAGMIGTTRPTH 799
Query: 298 YHILHDEIGFNPDDLQELVHSLSYVYQRSTTAISVVAPICYAHLAAAQVGQFIKFDEMXX 477
YH+L+DEIGF DDLQELVHSLSYVYQRSTTAISVVAP+CYAHLAAAQ+G +K++E+
Sbjct: 800 YHVLYDEIGFATDDLQELVHSLSYVYQRSTTAISVVAPVCYAHLAAAQMGTVMKYEELSE 859
Query: 478 XXXXXXXXXXXXXVPVQELPRLHEKVRSSMFFC 576
VPV +P+LH V +SMFFC
Sbjct: 860 TSSSHGGITTPGAVPVPPMPQLHNNVSTSMFFC 892
>sptr|Q9ZVD5|Q9ZVD5 Argonaute (AGO1)-like protein (Putative
post-transcriptional gene silencing protein).
Length = 924
Score = 229 bits (585), Expect = 3e-59
Identities = 102/153 (66%), Positives = 124/153 (81%)
Frame = +1
Query: 118 NPKFTLIIAQKNHHTKFFIPGKPDNVPAGTVVDNKVCHPRNFDFYMCSHAGMIGTTRPTH 297
NPKF L++AQKNHHTKFF P P+NVP GT++DNK+CHP+N DFY+C+HAGMIGTTRPTH
Sbjct: 772 NPKFLLLVAQKNHHTKFFQPTSPENVPPGTIIDNKICHPKNNDFYLCAHAGMIGTTRPTH 831
Query: 298 YHILHDEIGFNPDDLQELVHSLSYVYQRSTTAISVVAPICYAHLAAAQVGQFIKFDEMXX 477
YH+L+DEIGF+ D+LQELVHSLSYVYQRST+AISVVAPICYAHLAAAQ+G F+KF++
Sbjct: 832 YHVLYDEIGFSADELQELVHSLSYVYQRSTSAISVVAPICYAHLAAAQLGTFMKFEDQSE 891
Query: 478 XXXXXXXXXXXXXVPVQELPRLHEKVRSSMFFC 576
+ V +LPRL + V +SMFFC
Sbjct: 892 TSSSHGGITAPGPISVAQLPRLKDNVANSMFFC 924
>sptr|Q84YI4|Q84YI4 ARGONAUTE9 protein.
Length = 896
Score = 229 bits (585), Expect = 3e-59
Identities = 102/153 (66%), Positives = 124/153 (81%)
Frame = +1
Query: 118 NPKFTLIIAQKNHHTKFFIPGKPDNVPAGTVVDNKVCHPRNFDFYMCSHAGMIGTTRPTH 297
+PKFT+I+AQKNHHTKFF PDNVP GT++D+++CHPRNFDFY+C+HAGMIGTTRPTH
Sbjct: 744 HPKFTVIVAQKNHHTKFFQSRGPDNVPPGTIIDSQICHPRNFDFYLCAHAGMIGTTRPTH 803
Query: 298 YHILHDEIGFNPDDLQELVHSLSYVYQRSTTAISVVAPICYAHLAAAQVGQFIKFDEMXX 477
YH+L+DEIGF DDLQELVHSLS+VYQRSTTAISVVAP+CYAHLAAAQ+G +K++E+
Sbjct: 804 YHVLYDEIGFATDDLQELVHSLSHVYQRSTTAISVVAPVCYAHLAAAQMGTVMKYEELSE 863
Query: 478 XXXXXXXXXXXXXVPVQELPRLHEKVRSSMFFC 576
VPV +P+LH V +SMFFC
Sbjct: 864 TSSSHGGITTPGTVPVPPMPQLHNNVSTSMFFC 896
>sptrnew|BAC84805|BAC84805 Putative ARGONAUTE9 protein.
Length = 889
Score = 214 bits (545), Expect = 1e-54
Identities = 104/152 (68%), Positives = 116/152 (76%)
Frame = +1
Query: 121 PKFTLIIAQKNHHTKFFIPGKPDNVPAGTVVDNKVCHPRNFDFYMCSHAGMIGTTRPTHY 300
PKFT+IIAQKNHHTK F PDNVP GTVVD+ + HPR +DFYM +HAG IGT+RPTHY
Sbjct: 738 PKFTVIIAQKNHHTKLFQENTPDNVPPGTVVDSGIVHPRQYDFYMYAHAGPIGTSRPTHY 797
Query: 301 HILHDEIGFNPDDLQELVHSLSYVYQRSTTAISVVAPICYAHLAAAQVGQFIKFDEMXXX 480
H+L DEIGF PDD+Q+LV SLSYVYQRSTTAISVVAPICYAHLAAAQ+GQF+KF+E
Sbjct: 798 HVLLDEIGFLPDDVQKLVLSLSYVYQRSTTAISVVAPICYAHLAAAQMGQFMKFEEFAET 857
Query: 481 XXXXXXXXXXXXVPVQELPRLHEKVRSSMFFC 576
V ELPRLH V SSMFFC
Sbjct: 858 SSGSGGVPSSSGAVVPELPRLHADVCSSMFFC 889
>sptr|Q8S9N1|Q8S9N1 Zwille/pinhead-like protein (Fragment).
Length = 193
Score = 197 bits (502), Expect = 1e-49
Identities = 100/149 (67%), Positives = 112/149 (75%)
Frame = +1
Query: 121 PKFTLIIAQKNHHTKFFIPGKPDNVPAGTVVDNKVCHPRNFDFYMCSHAGMIGTTRPTHY 300
PKFT+IIAQKNHHTK F PDNVP GTVVD+ + HPR +DFYM +HAG IGT+RPTHY
Sbjct: 40 PKFTVIIAQKNHHTKLFQENTPDNVPPGTVVDSGIVHPRQYDFYMYAHAGPIGTSRPTHY 99
Query: 301 HILHDEIGFNPDDLQELVHSLSYVYQRSTTAISVVAPICYAHLAAAQVGQFIKFDEMXXX 480
H+L DEIGF PDD+Q+LV SLSYVYQRSTTAISVVAPICYAHLAAAQ+GQF+KF+E
Sbjct: 100 HVLLDEIGFLPDDVQKLVLSLSYVYQRSTTAISVVAPICYAHLAAAQMGQFMKFEEFAEL 159
Query: 481 XXXXXXXXXXXXVPVQELPRLHEKVRSSM 567
ELPRLH V SSM
Sbjct: 160 RLEVVVSFVIRS-SGPELPRLHADVCSSM 187
>sptr|Q7XBH5|Q7XBH5 Zwille pinhead-like protein.
Length = 361
Score = 191 bits (486), Expect = 9e-48
Identities = 90/116 (77%), Positives = 102/116 (87%)
Frame = +1
Query: 121 PKFTLIIAQKNHHTKFFIPGKPDNVPAGTVVDNKVCHPRNFDFYMCSHAGMIGTTRPTHY 300
PKFT+IIAQKNHHTK F PDNVP GTVVD+ + HPR +DFYM +HAG IGT+RPTHY
Sbjct: 212 PKFTVIIAQKNHHTKLFQENTPDNVPPGTVVDSGIVHPRQYDFYMYAHAGPIGTSRPTHY 271
Query: 301 HILHDEIGFNPDDLQELVHSLSYVYQRSTTAISVVAPICYAHLAAAQVGQFIKFDE 468
H+L DEIGF PDD+Q+LV SLSYVYQRSTTAISVVAPICYAHLAAAQ+GQF+KF+E
Sbjct: 272 HVLLDEIGFLPDDVQKLVLSLSYVYQRSTTAISVVAPICYAHLAAAQMGQFMKFEE 327
>sptr|O48771|O48771 Argonaute (AGO1)-like protein.
Length = 887
Score = 182 bits (462), Expect = 5e-45
Identities = 92/160 (57%), Positives = 107/160 (66%), Gaps = 8/160 (5%)
Frame = +1
Query: 121 PKFTLIIAQKNHHTKFFIPGKPDNVPAGTVVDNKVCHPRNFDFYMCSHAGMIGTTRPTHY 300
PKFT+I+AQKNHHTK F P+NVPAGTVVD K+ HP N+DFYMC+HAG IGT+RP HY
Sbjct: 740 PKFTVIVAQKNHHTKLFQAKGPENVPAGTVVDTKIVHPTNYDFYMCAHAGKIGTSRPAHY 799
Query: 301 HILHDEIGFNPDDLQELVHSLSYVYQRSTTAIS--------VVAPICYAHLAAAQVGQFI 456
H+L DEIGF+PDDLQ L+HSLSY S +S VAP+ YAHLAAAQV QF
Sbjct: 800 HVLLDEIGFSPDDLQNLIHSLSYKLLNSIFNVSSLLCVFVLSVAPVRYAHLAAAQVAQFT 859
Query: 457 KFDEMXXXXXXXXXXXXXXXVPVQELPRLHEKVRSSMFFC 576
KF+ + V ELPRLHE V +MFFC
Sbjct: 860 KFEGISEDG------------KVPELPRLHENVEGNMFFC 887
>sptr|Q8LP00|Q8LP00 ZLL/PNH homologous protein.
Length = 978
Score = 140 bits (354), Expect = 2e-32
Identities = 69/160 (43%), Positives = 91/160 (56%), Gaps = 8/160 (5%)
Frame = +1
Query: 121 PKFTLIIAQKNHHTKFFIPGKPD--------NVPAGTVVDNKVCHPRNFDFYMCSHAGMI 276
P T ++ QK HHT+ F D N+ GTVVD+K+CHP FDFY+CSHAG+
Sbjct: 820 PPVTFVVVQKRHHTRLFANNHKDRSSTDKSGNILPGTVVDSKICHPSEFDFYLCSHAGIQ 879
Query: 277 GTTRPTHYHILHDEIGFNPDDLQELVHSLSYVYQRSTTAISVVAPICYAHLAAAQVGQFI 456
GT+RP HYH+L DE F D++Q L ++L Y Y R T ++SVV P YAHLAA + +F
Sbjct: 880 GTSRPAHYHVLWDENNFTADEMQTLTNNLCYTYARCTRSVSVVPPAYYAHLAAFR-ARFY 938
Query: 457 KFDEMXXXXXXXXXXXXXXXVPVQELPRLHEKVRSSMFFC 576
EM V+ LP + EKV+ MF+C
Sbjct: 939 MEPEMSENQTTSKSSTGTNGTSVKPLPAVKEKVKRVMFYC 978
>sptr|Q851R2|Q851R2 Putative argonaute protein.
Length = 1058
Score = 140 bits (352), Expect = 3e-32
Identities = 69/163 (42%), Positives = 94/163 (57%), Gaps = 11/163 (6%)
Frame = +1
Query: 121 PKFTLIIAQKNHHTKFF--------IPGKPDNVPAGTVVDNKVCHPRNFDFYMCSHAGMI 276
P T ++ QK HHT+ F + K N+ GTVVD ++CHP FDFY+CSHAG+
Sbjct: 896 PPVTFVVVQKRHHTRLFPEVHGRRDMTDKSGNILPGTVVDRQICHPTEFDFYLCSHAGIQ 955
Query: 277 GTTRPTHYHILHDEIGFNPDDLQELVHSLSYVYQRSTTAISVVAPICYAHLAAAQVGQFI 456
GT+RPTHYH+L+DE F D LQ L ++L Y Y R T A+SVV P YAHLAA + ++
Sbjct: 956 GTSRPTHYHVLYDENHFTADALQSLTNNLCYTYARCTRAVSVVPPAYYAHLAAFRARYYV 1015
Query: 457 K---FDEMXXXXXXXXXXXXXXXVPVQELPRLHEKVRSSMFFC 576
+ D V V++LP++ E V+ MF+C
Sbjct: 1016 EGESSDGGSTPGSSGQAVAREGPVEVRQLPKIKENVKDVMFYC 1058
>sptrnew|BAC83909|BAC83909 Putative leaf development protein
Argonaute.
Length = 1052
Score = 137 bits (345), Expect = 2e-31
Identities = 70/164 (42%), Positives = 93/164 (56%), Gaps = 12/164 (7%)
Frame = +1
Query: 121 PKFTLIIAQKNHHTKFF--IPGKPD------NVPAGTVVDNKVCHPRNFDFYMCSHAGMI 276
P T ++ QK HHT+ F + G+ D NV GTVVD +CHP FDFY+CSHAG+
Sbjct: 889 PLVTFVVVQKRHHTRLFPEVHGRQDLTDRSGNVRPGTVVDTNICHPSEFDFYLCSHAGIQ 948
Query: 277 GTTRPTHYHILHDEIGFNPDDLQELVHSLSYVYQRSTTAISVVAPICYAHLAAAQVGQFI 456
GT+RPTHYH+LHDE F+ D LQ L ++L Y Y R T ++SVV P YAHLAA + +
Sbjct: 949 GTSRPTHYHVLHDENRFSADQLQMLTYNLCYTYARCTRSVSVVPPAYYAHLAAFRARYYD 1008
Query: 457 KFDEMXXXXXXXXXXXXXXX----VPVQELPRLHEKVRSSMFFC 576
+ M V+ LP++ E V+ MF+C
Sbjct: 1009 EPPAMDGASSVGSGGNQAAAGGQPPAVRRLPQIKENVKDVMFYC 1052
>sw|Q9SJK3|AGOL_ARATH Argonaute-like protein At2g27880.
Length = 997
Score = 136 bits (343), Expect = 3e-31
Identities = 64/160 (40%), Positives = 90/160 (56%), Gaps = 8/160 (5%)
Frame = +1
Query: 121 PKFTLIIAQKNHHTKFF--------IPGKPDNVPAGTVVDNKVCHPRNFDFYMCSHAGMI 276
P+ T +I QK HHT+ F + K N+ GTVVD K+CHP FDFY+ SHAG+
Sbjct: 838 PRVTFVIVQKRHHTRLFPEQHGNRDMTDKSGNIQPGTVVDTKICHPNEFDFYLNSHAGIQ 897
Query: 277 GTTRPTHYHILHDEIGFNPDDLQELVHSLSYVYQRSTTAISVVAPICYAHLAAAQVGQFI 456
GT+RP HYH+L DE GF D LQ L ++L Y Y R T ++S+V P YAHLAA + ++
Sbjct: 898 GTSRPAHYHVLLDENGFTADQLQMLTNNLCYTYARCTKSVSIVPPAYYAHLAAFRARYYM 957
Query: 457 KFDEMXXXXXXXXXXXXXXXVPVQELPRLHEKVRSSMFFC 576
+ + + +LP + + V+ MF+C
Sbjct: 958 ESEMSDGGSSRSRSSTTGVGQVISQLPAIKDNVKEVMFYC 997
>sptr|Q7Y001|Q7Y001 Putative leaf development and shoot apical
meristem regulating protein.
Length = 1055
Score = 136 bits (342), Expect = 4e-31
Identities = 65/158 (41%), Positives = 89/158 (56%), Gaps = 6/158 (3%)
Frame = +1
Query: 121 PKFTLIIAQKNHHTKFFIPGKPD------NVPAGTVVDNKVCHPRNFDFYMCSHAGMIGT 282
P T ++ QK HHT+ F + D N+ GTVVD +CHP FDFY+CSH+G+ GT
Sbjct: 900 PPVTFVVVQKRHHTRLFPENRRDMMDRSGNILPGTVVDTMICHPSEFDFYLCSHSGIKGT 959
Query: 283 TRPTHYHILHDEIGFNPDDLQELVHSLSYVYQRSTTAISVVAPICYAHLAAAQVGQFIKF 462
+RPTHYH+L DE GF D LQ L ++LSY Y R T A+S+V P YAHL A + +++
Sbjct: 960 SRPTHYHVLLDENGFKADTLQTLTYNLSYTYARCTRAVSIVPPAYYAHLGAFRARYYMED 1019
Query: 463 DEMXXXXXXXXXXXXXXXVPVQELPRLHEKVRSSMFFC 576
+ + LP + E V+ MF+C
Sbjct: 1020 EHSDQGSSSSVTTRTDRS--TKPLPEIKENVKRFMFYC 1055
>sptr|Q7XZZ9|Q7XZZ9 Putative leaf development and shoot apical
meristem regulating protein.
Length = 895
Score = 134 bits (338), Expect = 1e-30
Identities = 64/165 (38%), Positives = 92/165 (55%), Gaps = 13/165 (7%)
Frame = +1
Query: 121 PKFTLIIAQKNHHTKFFIPG--------KPDNVPAGTVVDNKVCHPRNFDFYMCSHAGMI 276
P T ++ QK HHT+ F + N+ GTVVD K+CHP FDFY+CSH+G+
Sbjct: 731 PPVTFVVVQKRHHTRLFPEDHHARDQMDRSRNILPGTVVDTKICHPSEFDFYLCSHSGIQ 790
Query: 277 GTTRPTHYHILHDEIGFNPDDLQELVHSLSYVYQRSTTAISVVAPICYAHLAAAQVGQFI 456
GT+ PTHY++L DE F+ D LQ L + L Y Y R T ++S+V P+ YAHLAA++ ++
Sbjct: 791 GTSHPTHYYVLFDENNFSADALQTLTYHLCYTYARCTRSVSIVPPVYYAHLAASRARHYL 850
Query: 457 KFDEM-----XXXXXXXXXXXXXXXVPVQELPRLHEKVRSSMFFC 576
+ + VPV+ LP + E V+ MF+C
Sbjct: 851 EEGSLPDHGSSSASAAGGSRRNDRGVPVKPLPEIKENVKQFMFYC 895
>sw|Q9XGW1|PINH_ARATH PINHEAD protein (ZWILLE protein).
Length = 988
Score = 134 bits (336), Expect = 2e-30
Identities = 68/163 (41%), Positives = 90/163 (55%), Gaps = 11/163 (6%)
Frame = +1
Query: 121 PKFTLIIAQKNHHTKFFIPGKPD--------NVPAGTVVDNKVCHPRNFDFYMCSHAGMI 276
P T I+ QK HHT+ F D N+ GTVVD K+CHP FDFY+CSHAG+
Sbjct: 826 PPVTFIVVQKRHHTRLFANNHRDKNSTDRSGNILPGTVVDTKICHPTEFDFYLCSHAGIQ 885
Query: 277 GTTRPTHYHILHDEIGFNPDDLQELVHSLSYVYQRSTTAISVVAPICYAHLAAAQVGQFI 456
GT+RP HYH+L DE F D +Q L ++L Y Y R T ++S+V P YAHLAA + ++
Sbjct: 886 GTSRPAHYHVLWDENNFTADGIQSLTNNLCYTYARCTRSVSIVPPAYYAHLAAFRARFYL 945
Query: 457 KFDEMXXXXX---XXXXXXXXXXVPVQELPRLHEKVRSSMFFC 576
+ + M V V+ LP L E V+ MF+C
Sbjct: 946 EPEIMQDNGSPGKKNTKTTTVGDVGVKPLPALKENVKRVMFYC 988
>sptrnew|AAQ92355|AAQ92355 ZIPPY.
Length = 990
Score = 133 bits (334), Expect = 4e-30
Identities = 67/157 (42%), Positives = 90/157 (57%), Gaps = 2/157 (1%)
Frame = +1
Query: 112 EMNPKFTLIIAQKNHHTKFF-IPGKPDNVPAGTVVDNKVCHPRNFDFYMCSHAGMIGTTR 288
+ NP T + QK HHT+ F +N+P GTVVD + HP+ FDFY+CSH G+ GT+R
Sbjct: 834 DYNPSITFAVVQKRHHTRLFRCDPDHENIPPGTVVDTVITHPKEFDFYLCSHLGVKGTSR 893
Query: 289 PTHYHILHDEIGFNPDDLQELVHSLSYVYQRSTTAISVVAPICYAHLAAAQVGQFIKFDE 468
PTHYHIL DE F D+LQ LV++L Y + R T IS+V P YAHLAA + +I+
Sbjct: 894 PTHYHILWDENEFTSDELQRLVYNLCYTFVRCTKPISIVPPAYYAHLAAYRGRLYIERSS 953
Query: 469 MXXXXXXXXXXXXXXXVP-VQELPRLHEKVRSSMFFC 576
P LP+L + V++ MF+C
Sbjct: 954 ESNGGSMNPSSVSRVGPPKTIPLPKLSDNVKNLMFYC 990
>sptr|Q9C793|Q9C793 Pinhead-like protein.
Length = 990
Score = 133 bits (334), Expect = 4e-30
Identities = 67/157 (42%), Positives = 90/157 (57%), Gaps = 2/157 (1%)
Frame = +1
Query: 112 EMNPKFTLIIAQKNHHTKFF-IPGKPDNVPAGTVVDNKVCHPRNFDFYMCSHAGMIGTTR 288
+ NP T + QK HHT+ F +N+P GTVVD + HP+ FDFY+CSH G+ GT+R
Sbjct: 834 DYNPSITFAVVQKRHHTRLFRCDPDHENIPPGTVVDTVITHPKEFDFYLCSHLGVKGTSR 893
Query: 289 PTHYHILHDEIGFNPDDLQELVHSLSYVYQRSTTAISVVAPICYAHLAAAQVGQFIKFDE 468
PTHYHIL DE F D+LQ LV++L Y + R T IS+V P YAHLAA + +I+
Sbjct: 894 PTHYHILWDENEFTSDELQRLVYNLCYTFVRCTKPISIVPPAYYAHLAAYRGRLYIERSS 953
Query: 469 MXXXXXXXXXXXXXXXVP-VQELPRLHEKVRSSMFFC 576
P LP+L + V++ MF+C
Sbjct: 954 ESNGGSMNPSSVSRVGPPKTIPLPKLSDNVKNLMFYC 990
>sptrnew|AAN41341|AAN41341 Putative leaf development protein
Argonaute.
Length = 1048
Score = 129 bits (324), Expect = 5e-29
Identities = 66/172 (38%), Positives = 90/172 (52%), Gaps = 20/172 (11%)
Frame = +1
Query: 121 PKFTLIIAQKNHHTKFFIPGKPD--------NVPAGTVVDNKVCHPRNFDFYMCSHAGMI 276
P T ++ QK HHT+ F D N+ GTVVD+K+CHP FDFY+CSHAG+
Sbjct: 877 PPVTFVVVQKRHHTRLFAQNHNDRHSVDRSGNILPGTVVDSKICHPTEFDFYLCSHAGIQ 936
Query: 277 GTTRPTHYHILHDEIGFNPDDLQELVHSLSYVYQRSTTAISVVAPICYAHLAAAQVGQFI 456
GT+RP HYH+L DE F D LQ L ++L Y Y R T ++S+V P YAHLAA + ++
Sbjct: 937 GTSRPAHYHVLWDENNFTADGLQSLTNNLCYTYARCTRSVSIVPPAYYAHLAAFRARFYM 996
Query: 457 KFDEMXXXXXXXXXXXXXXXV------------PVQELPRLHEKVRSSMFFC 576
+ + + V+ LP L E V+ MF+C
Sbjct: 997 EPETSDSGSMASGSMARGGGMAGRSTRGPNVNAAVRPLPALKENVKRVMFYC 1048
>sw|O04379|AGO1_ARATH Argonaute protein.
Length = 1048
Score = 129 bits (324), Expect = 5e-29
Identities = 66/172 (38%), Positives = 90/172 (52%), Gaps = 20/172 (11%)
Frame = +1
Query: 121 PKFTLIIAQKNHHTKFFIPGKPD--------NVPAGTVVDNKVCHPRNFDFYMCSHAGMI 276
P T ++ QK HHT+ F D N+ GTVVD+K+CHP FDFY+CSHAG+
Sbjct: 877 PPVTFVVVQKRHHTRLFAQNHNDRHSVDRSGNILPGTVVDSKICHPTEFDFYLCSHAGIQ 936
Query: 277 GTTRPTHYHILHDEIGFNPDDLQELVHSLSYVYQRSTTAISVVAPICYAHLAAAQVGQFI 456
GT+RP HYH+L DE F D LQ L ++L Y Y R T ++S+V P YAHLAA + ++
Sbjct: 937 GTSRPAHYHVLWDENNFTADGLQSLTNNLCYTYARCTRSVSIVPPAYYAHLAAFRARFYM 996
Query: 457 KFDEMXXXXXXXXXXXXXXXV------------PVQELPRLHEKVRSSMFFC 576
+ + + V+ LP L E V+ MF+C
Sbjct: 997 EPETSDSGSMASGSMARGGGMAGRSTRGPNVNAAVRPLPALKENVKRVMFYC 1048
>sptr|Q8LNZ9|Q8LNZ9 AGO1 homologous protein.
Length = 909
Score = 128 bits (322), Expect = 9e-29
Identities = 67/173 (38%), Positives = 90/173 (52%), Gaps = 21/173 (12%)
Frame = +1
Query: 121 PKFTLIIAQKNHHTKFFIPGKPD--------NVPAGTVVDNKVCHPRNFDFYMCSHAGMI 276
PK T I+ QK HHT+ F D N+ GTVVD+K+CHP FDFY+CSHAG+
Sbjct: 737 PKVTFIVVQKRHHTRLFAHNHNDQNSVDRSGNILPGTVVDSKICHPTEFDFYLCSHAGIK 796
Query: 277 GTTRPTHYHILHDEIGFNPDDLQELVHSLSYVYQRSTTAISVVAPICYAHLAAAQVGQFI 456
GT+RP HYH+L +E F D LQ L ++L Y Y R T ++S+V P YAHLAA + ++
Sbjct: 797 GTSRPAHYHVLWNENNFTADALQILTNNLCYTYARCTRSVSIVPPAYYAHLAAFRARFYM 856
Query: 457 KFDEMXXXX-------------XXXXXXXXXXXVPVQELPRLHEKVRSSMFFC 576
+ D V+ LP L + V+ MF+C
Sbjct: 857 EPDTSDSSSVVSGPGVRGPLSGSSTSRTRAPGGAAVKPLPALKDSVKRVMFYC 909
>sptr|Q7XSA2|Q7XSA2 OSJNBa0005N02.3 protein.
Length = 1254
Score = 124 bits (312), Expect = 1e-27
Identities = 57/113 (50%), Positives = 73/113 (64%), Gaps = 8/113 (7%)
Frame = +1
Query: 121 PKFTLIIAQKNHHTKFFIPGKPD--------NVPAGTVVDNKVCHPRNFDFYMCSHAGMI 276
P T ++ QK HHT+ F D N+ GTVVD+K+CHP FDFY+CSHAG+
Sbjct: 1033 PPVTFVVVQKRHHTRLFANNHNDQRTVDRSGNILPGTVVDSKICHPTEFDFYLCSHAGIQ 1092
Query: 277 GTTRPTHYHILHDEIGFNPDDLQELVHSLSYVYQRSTTAISVVAPICYAHLAA 435
GT+RP HYH+L DE F D+LQ L ++L Y Y R T ++S+V P YAHLAA
Sbjct: 1093 GTSRPAHYHVLWDENKFTADELQTLTNNLCYTYARCTRSVSIVPPAYYAHLAA 1145
>sw|Q9QZ81|I2C2_RAT Eukaryotic translation initiation factor 2C 2
(eIF2C 2) (eIF-2C 2) (Golgi ER protein 95 kDa) (GERp95).
Length = 863
Score = 124 bits (312), Expect = 1e-27
Identities = 57/114 (50%), Positives = 74/114 (64%), Gaps = 6/114 (5%)
Frame = +1
Query: 112 EMNPKFTLIIAQKNHHTKFFIP------GKPDNVPAGTVVDNKVCHPRNFDFYMCSHAGM 273
E P T I+ QK HHT+ F GK N+PAGT VD K+ HP FDFY+CSHAG+
Sbjct: 701 EYQPGITFIVVQKRHHTRLFCTDKNERVGKSGNIPAGTTVDTKITHPTEFDFYLCSHAGI 760
Query: 274 IGTTRPTHYHILHDEIGFNPDDLQELVHSLSYVYQRSTTAISVVAPICYAHLAA 435
GT+RP+HYH+L D+ F+ D+LQ L + L + Y R T ++S+ AP YAHL A
Sbjct: 761 QGTSRPSHYHVLWDDNRFSSDELQILTYQLCHTYVRCTRSVSIPAPAYYAHLVA 814
>sptr|Q9LP83|Q9LP83 T1N15.2.
Length = 1123
Score = 124 bits (310), Expect = 2e-27
Identities = 66/175 (37%), Positives = 90/175 (51%), Gaps = 23/175 (13%)
Frame = +1
Query: 121 PKFTLIIAQKNHHTKFFIPGKPD--------NVPAG---TVVDNKVCHPRNFDFYMCSHA 267
P T ++ QK HHT+ F D N+ G TVVD+K+CHP FDFY+CSHA
Sbjct: 949 PPVTFVVVQKRHHTRLFAQNHNDRHSVDRSGNILPGETSTVVDSKICHPTEFDFYLCSHA 1008
Query: 268 GMIGTTRPTHYHILHDEIGFNPDDLQELVHSLSYVYQRSTTAISVVAPICYAHLAAAQVG 447
G+ GT+RP HYH+L DE F D LQ L ++L Y Y R T ++S+V P YAHLAA +
Sbjct: 1009 GIQGTSRPAHYHVLWDENNFTADGLQSLTNNLCYTYARCTRSVSIVPPAYYAHLAAFRAR 1068
Query: 448 QFIKFDEMXXXXXXXXXXXXXXXV------------PVQELPRLHEKVRSSMFFC 576
+++ + + V+ LP L E V+ MF+C
Sbjct: 1069 FYMEPETSDSGSMASGSMARGGGMAGRSTRGPNVNAAVRPLPALKENVKRVMFYC 1123
>sptr|Q852N2|Q852N2 Putative argonaute protein.
Length = 1192
Score = 123 bits (309), Expect = 3e-27
Identities = 58/121 (47%), Positives = 77/121 (63%), Gaps = 8/121 (6%)
Frame = +1
Query: 121 PKFTLIIAQKNHHTKFF--------IPGKPDNVPAGTVVDNKVCHPRNFDFYMCSHAGMI 276
P T ++ QK HHT+ F + K N+ GTV D ++CHP F FY+CSHAG+
Sbjct: 650 PPVTFVVVQKRHHTRLFPEVHGRRDMTDKSGNILPGTVKDRQICHPTEFYFYLCSHAGIQ 709
Query: 277 GTTRPTHYHILHDEIGFNPDDLQELVHSLSYVYQRSTTAISVVAPICYAHLAAAQVGQFI 456
GT+RPTHYH+L+DE F D+LQ L ++L Y+Y R T A+SVV P Y+HLAA+ I
Sbjct: 710 GTSRPTHYHVLYDENHFTADELQTLTNNLCYIYARCTHAVSVVPPAYYSHLAASHAHCCI 769
Query: 457 K 459
K
Sbjct: 770 K 770
>sw|O77503|I2C2_RABIT Eukaryotic translation initiation factor 2C 2
(eIF2C 2) (eIF-2C 2).
Length = 813
Score = 123 bits (309), Expect = 3e-27
Identities = 56/114 (49%), Positives = 74/114 (64%), Gaps = 6/114 (5%)
Frame = +1
Query: 112 EMNPKFTLIIAQKNHHTKFFIP------GKPDNVPAGTVVDNKVCHPRNFDFYMCSHAGM 273
+ P T I+ QK HHT+ F GK N+PAGT VD K+ HP FDFY+CSHAG+
Sbjct: 651 DYQPGITFIVVQKRHHTRLFCTDKNERVGKSGNIPAGTTVDTKITHPTEFDFYLCSHAGI 710
Query: 274 IGTTRPTHYHILHDEIGFNPDDLQELVHSLSYVYQRSTTAISVVAPICYAHLAA 435
GT+RP+HYH+L D+ F+ D+LQ L + L + Y R T ++S+ AP YAHL A
Sbjct: 711 QGTSRPSHYHVLWDDNRFSSDELQILTYQLCHTYVRCTRSVSIPAPAYYAHLVA 764
>sptr|Q8TCZ5|Q8TCZ5 Eukaryotic initiation factor 2C2.
Length = 851
Score = 123 bits (309), Expect = 3e-27
Identities = 56/114 (49%), Positives = 74/114 (64%), Gaps = 6/114 (5%)
Frame = +1
Query: 112 EMNPKFTLIIAQKNHHTKFFIP------GKPDNVPAGTVVDNKVCHPRNFDFYMCSHAGM 273
+ P T I+ QK HHT+ F GK N+PAGT VD K+ HP FDFY+CSHAG+
Sbjct: 689 DYQPGITFIVVQKRHHTRLFCTDKNERVGKSGNIPAGTTVDTKITHPTEFDFYLCSHAGI 748
Query: 274 IGTTRPTHYHILHDEIGFNPDDLQELVHSLSYVYQRSTTAISVVAPICYAHLAA 435
GT+RP+HYH+L D+ F+ D+LQ L + L + Y R T ++S+ AP YAHL A
Sbjct: 749 QGTSRPSHYHVLWDDNRFSSDELQILTYQLCHTYVRCTRSVSIPAPAYYAHLVA 802
>sptr|Q96ID1|Q96ID1 Hypothetical protein.
Length = 377
Score = 123 bits (309), Expect = 3e-27
Identities = 56/114 (49%), Positives = 74/114 (64%), Gaps = 6/114 (5%)
Frame = +1
Query: 112 EMNPKFTLIIAQKNHHTKFFIP------GKPDNVPAGTVVDNKVCHPRNFDFYMCSHAGM 273
+ P T I+ QK HHT+ F GK N+PAGT VD K+ HP FDFY+CSHAG+
Sbjct: 215 DYQPGITFIVVQKRHHTRLFCTDKNERVGKSGNIPAGTTVDTKITHPTEFDFYLCSHAGI 274
Query: 274 IGTTRPTHYHILHDEIGFNPDDLQELVHSLSYVYQRSTTAISVVAPICYAHLAA 435
GT+RP+HYH+L D+ F+ D+LQ L + L + Y R T ++S+ AP YAHL A
Sbjct: 275 QGTSRPSHYHVLWDDNRFSSDELQILTYQLCHTYVRCTRSVSIPAPAYYAHLVA 328
>sw|Q9UKV8|I2C2_HUMAN Eukaryotic translation initiation factor 2C 2
(eIF2C 2) (eIF-2C 2) (Fragment).
Length = 377
Score = 123 bits (309), Expect = 3e-27
Identities = 56/114 (49%), Positives = 74/114 (64%), Gaps = 6/114 (5%)
Frame = +1
Query: 112 EMNPKFTLIIAQKNHHTKFFIP------GKPDNVPAGTVVDNKVCHPRNFDFYMCSHAGM 273
+ P T I+ QK HHT+ F GK N+PAGT VD K+ HP FDFY+CSHAG+
Sbjct: 215 DYQPGITFIVVQKRHHTRLFCTDKNERVGKSGNIPAGTTVDTKITHPTEFDFYLCSHAGI 274
Query: 274 IGTTRPTHYHILHDEIGFNPDDLQELVHSLSYVYQRSTTAISVVAPICYAHLAA 435
GT+RP+HYH+L D+ F+ D+LQ L + L + Y R T ++S+ AP YAHL A
Sbjct: 275 QGTSRPSHYHVLWDDNRFSSDELQILTYQLCHTYVRCTRSVSIPAPAYYAHLVA 328
>sptrnew|AAH56639|AAH56639 Hypothetical protein (Fragment).
Length = 437
Score = 123 bits (309), Expect = 3e-27
Identities = 56/114 (49%), Positives = 74/114 (64%), Gaps = 6/114 (5%)
Frame = +1
Query: 112 EMNPKFTLIIAQKNHHTKFFIP------GKPDNVPAGTVVDNKVCHPRNFDFYMCSHAGM 273
+ P T I+ QK HHT+ F GK N+PAGT VD K+ HP FDFY+CSHAG+
Sbjct: 275 DYQPGITFIVVQKRHHTRLFCTDKNERVGKSGNIPAGTTVDTKITHPTEFDFYLCSHAGI 334
Query: 274 IGTTRPTHYHILHDEIGFNPDDLQELVHSLSYVYQRSTTAISVVAPICYAHLAA 435
GT+RP+HYH+L D+ F+ D+LQ L + L + Y R T ++S+ AP YAHL A
Sbjct: 335 QGTSRPSHYHVLWDDNRFSSDELQILTYQLCHTYVRCTRSVSIPAPAYYAHLVA 388
>sptr|Q8R3Q7|Q8R3Q7 Similar to eukaryotic translation initiation
factor 2C, 2 (Fragment).
Length = 530
Score = 123 bits (309), Expect = 3e-27
Identities = 56/114 (49%), Positives = 74/114 (64%), Gaps = 6/114 (5%)
Frame = +1
Query: 112 EMNPKFTLIIAQKNHHTKFFIP------GKPDNVPAGTVVDNKVCHPRNFDFYMCSHAGM 273
+ P T I+ QK HHT+ F GK N+PAGT VD K+ HP FDFY+CSHAG+
Sbjct: 368 DYQPGITFIVVQKRHHTRLFCTDKNERVGKSGNIPAGTTVDTKITHPTEFDFYLCSHAGI 427
Query: 274 IGTTRPTHYHILHDEIGFNPDDLQELVHSLSYVYQRSTTAISVVAPICYAHLAA 435
GT+RP+HYH+L D+ F+ D+LQ L + L + Y R T ++S+ AP YAHL A
Sbjct: 428 QGTSRPSHYHVLWDDNRFSSDELQILTYQLCHTYVRCTRSVSIPAPAYYAHLVA 481
Database: /db/trembl-ebi/tmp/swall
Posted date: Oct 24, 2003 8:22 PM
Number of letters in database: 411,996,502
Number of sequences in database: 1,288,562
Lambda K H
0.318 0.135 0.401
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 672,934,253
Number of Sequences: 1288562
Number of extensions: 15052919
Number of successful extensions: 37573
Number of sequences better than 10.0: 30
Number of HSP's better than 10.0 without gapping: 36121
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 37481
length of database: 411,996,502
effective HSP length: 121
effective length of database: 256,080,500
effective search space used: 42765443500
frameshift window, decay const: 50, 0.1
T: 12
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)

