BLASTX 2.2.6 [Apr-09-2003]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
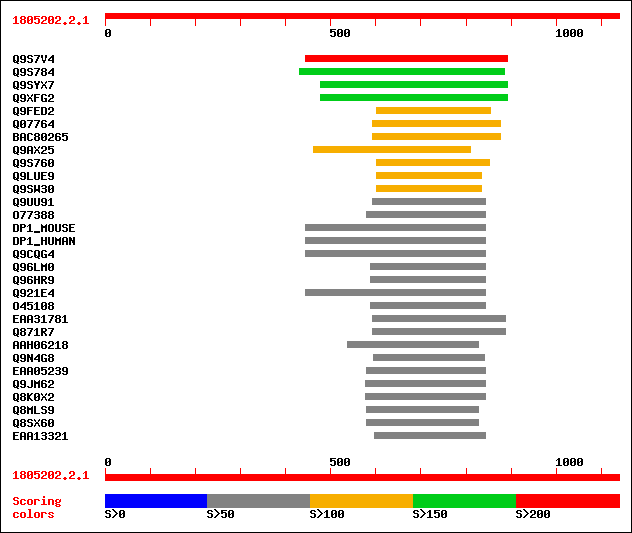
Query= 1805202.2.1
(1139 letters)
Database: /db/trembl-ebi/tmp/swall
1,272,877 sequences; 406,886,216 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
sptr|Q9S7V4|Q9S7V4 ATHVA22A. 212 7e-54
sptr|Q9S784|Q9S784 ATHVA22C. 186 4e-46
sptr|Q9SYX7|Q9SYX7 AtHVA22b. 161 1e-38
sptr|Q9XFG2|Q9XFG2 ATHVA22B. 161 2e-38
sptr|Q9FED2|Q9FED2 AtHVA22e (Abscisic acid-induced-like protein). 118 1e-25
sptr|Q07764|Q07764 Abscisic acid and stress inducible (A22) gene. 115 9e-25
sptrnew|BAC80265|BAC80265 Hypothetical protein Wrab15. 115 1e-24
sptr|Q9AX25|Q9AX25 P0456A01.10 protein (P0042A10.34 protein). 114 3e-24
sptr|Q9S760|Q9S760 ATHVA22D (Putative abscisic acid-induced prot... 110 3e-23
sptr|Q9LUE9|Q9LUE9 Genomic DNA, chromosome 5, P1 clone:MFB16. 108 1e-22
sptr|Q9SW30|Q9SW30 Abscisic acid-induced-like protein. 102 1e-20
sptr|Q9UU91|Q9UU91 Putative transport protein. 79 1e-13
sptr|O77388|O77388 Hypothetical 26.0 kDa protein. 78 2e-13
sw|Q60870|DP1_MOUSE Polyposis locus protein 1 homolog (TB2 prote... 77 6e-13
sw|Q00765|DP1_HUMAN Polyposis locus protein 1 (TB2 protein). 75 2e-12
sptr|Q9CQG4|Q9CQG4 Deleted in polyposis 1. 74 3e-12
sptr|Q96LM0|Q96LM0 Hypothetical protein FLJ25383 (TB2 protein-li... 74 5e-12
sptr|Q96HR9|Q96HR9 Hypothetical protein. 74 5e-12
sptr|Q921E4|Q921E4 Deleted in polyposis 1. 73 9e-12
sptr|O45108|O45108 Hypothetical 22.8 kDa protein. 72 1e-11
sptrnew|EAA31781|EAA31781 Hypothetical protein. 72 1e-11
sptr|Q871R7|Q871R7 Related to Ypt-interacting protein YIP2. 72 1e-11
sptrnew|AAH06218|AAH06218 Hypothetical protein. 72 1e-11
sptr|Q9N4G8|Q9N4G8 Hypothetical 20.6 kDa protein. 72 1e-11
sptrnew|EAA05239|EAA05239 ENSANGP00000018512 (Fragment). 71 3e-11
sptr|Q9JM62|Q9JM62 Polyposis locus protein 1-like 1 (TB2 protein... 70 6e-11
sptr|Q8K0X2|Q8K0X2 Hypothetical protein. 70 6e-11
sptr|Q8MLS9|Q8MLS9 CG30193-PA (RE29641p). 69 1e-10
sptr|Q8SX60|Q8SX60 LD42159p (CG30193-PD). 69 1e-10
sptrnew|EAA13321|EAA13321 AgCP2771 (Fragment). 69 2e-10
>sptr|Q9S7V4|Q9S7V4 ATHVA22A.
Length = 177
Score = 212 bits (540), Expect = 7e-54
Identities = 96/150 (64%), Positives = 122/150 (81%), Gaps = 1/150 (0%)
Frame = -3
Query: 891 SFLKVLVNNMDVLAGPLVSLAYPLYASVRAIETKSAVDDQQWLTYWVLYSFITLFELTFA 712
+FLKVL+ N DVLAGP+VSL YPLYASV+AIET+S DD+QWLTYWVLYS +TL ELTFA
Sbjct: 7 NFLKVLLRNFDVLAGPVVSLVYPLYASVQAIETQSHADDKQWLTYWVLYSLLTLIELTFA 66
Query: 711 PVLEWLPFWSYAKLFFNCWLVLPQFNGAAHVYEHFVRPMIVNQQVVNIWYIPKK-DESSR 535
++EWLP WSY KL CWLV+P F+GAA+VYEHFVRP+ VN + +NIWY+PKK D +
Sbjct: 67 KLIEWLPIWSYMKLILTCWLVIPYFSGAAYVYEHFVRPVFVNPRSINIWYVPKKMDIFRK 126
Query: 534 PDDVISAAQRYIEQNGSKAFENLVNKFKSS 445
PDDV++AA++YI +NG AFE ++++ S
Sbjct: 127 PDDVLTAAEKYIAENGPDAFEKILSRADKS 156
>sptr|Q9S784|Q9S784 ATHVA22C.
Length = 184
Score = 186 bits (473), Expect = 4e-46
Identities = 89/158 (56%), Positives = 116/158 (73%), Gaps = 6/158 (3%)
Frame = -3
Query: 885 LKVLVNNMDVLAGPLVSLAYPLYASVRAIETKSAVDDQQWLTYWVLYSFITLFELTFAPV 706
L+VL+ N DVLA PLV+L YPLYASV+AIET+S +D+QWLTYWVLY+ I+LFELTF+
Sbjct: 11 LQVLIKNFDVLALPLVTLVYPLYASVKAIETRSLPEDEQWLTYWVLYALISLFELTFSKP 70
Query: 705 LEWLPFWSYAKLFFNCWLVLPQFNGAAHVYEHFVRPMIVNQQ--VVNIWYIPKKDESSRP 532
LEW P W Y KLF CWLVLPQFNGA H+Y+HF+RP + Q IWY+P K + P
Sbjct: 71 LEWFPIWPYMKLFGICWLVLPQFNGAEHIYKHFIRPFYRDPQRATTKIWYVPHKKFNFFP 130
Query: 531 ----DDVISAAQRYIEQNGSKAFENLVNKFKSSNPRRS 430
DD+++AA++Y+EQ+G++AFE ++ K S RS
Sbjct: 131 KRDDDDILTAAEKYMEQHGTEAFERMIVKKDSYERGRS 168
>sptr|Q9SYX7|Q9SYX7 AtHVA22b.
Length = 167
Score = 161 bits (408), Expect = 1e-38
Identities = 75/138 (54%), Positives = 99/138 (71%)
Frame = -3
Query: 891 SFLKVLVNNMDVLAGPLVSLAYPLYASVRAIETKSAVDDQQWLTYWVLYSFITLFELTFA 712
S +KV+ N DV+AGP++SL YPLYASVRAIE++S DD+QWLTYW LYS I LFELTF
Sbjct: 7 SLVKVIFKNFDVIAGPMISLVYPLYASVRAIESRSHGDDKQWLTYWALYSLIKLFELTFF 66
Query: 711 PVLEWLPFWSYAKLFFNCWLVLPQFNGAAHVYEHFVRPMIVNQQVVNIWYIPKKDESSRP 532
+LEW+P + YAKL WLVLP NGAA++YEH+VR +++ VN+WY+P K +
Sbjct: 67 RLLEWIPLYPYAKLALTSWLVLPGMNGAAYLYEHYVRSFLLSPHTVNVWYVPAK----KD 122
Query: 531 DDVISAAQRYIEQNGSKA 478
DD+ + A ++ N S A
Sbjct: 123 DDLGATAGKFTPVNDSGA 140
>sptr|Q9XFG2|Q9XFG2 ATHVA22B.
Length = 167
Score = 161 bits (407), Expect = 2e-38
Identities = 75/138 (54%), Positives = 99/138 (71%)
Frame = -3
Query: 891 SFLKVLVNNMDVLAGPLVSLAYPLYASVRAIETKSAVDDQQWLTYWVLYSFITLFELTFA 712
S +KV+ N DV+AGP++SL YPLYASVRAIE++S DD+QWLTYW LYS I LFELTF
Sbjct: 7 SLVKVIFKNFDVIAGPVISLVYPLYASVRAIESRSHGDDKQWLTYWALYSLIKLFELTFF 66
Query: 711 PVLEWLPFWSYAKLFFNCWLVLPQFNGAAHVYEHFVRPMIVNQQVVNIWYIPKKDESSRP 532
+LEW+P + YAKL WLVLP NGAA++YEH+VR +++ VN+WY+P K +
Sbjct: 67 RLLEWIPLYPYAKLALTSWLVLPGMNGAAYLYEHYVRSFLLSPHTVNVWYVPAK----KD 122
Query: 531 DDVISAAQRYIEQNGSKA 478
DD+ + A ++ N S A
Sbjct: 123 DDLGATAGKFTPVNDSGA 140
>sptr|Q9FED2|Q9FED2 AtHVA22e (Abscisic acid-induced-like protein).
Length = 116
Score = 118 bits (296), Expect = 1e-25
Identities = 55/85 (64%), Positives = 63/85 (74%)
Frame = -3
Query: 855 LAGPLVSLAYPLYASVRAIETKSAVDDQQWLTYWVLYSFITLFELTFAPVLEWLPFWSYA 676
LAGP+V L YPLYASV AIE+ S VDD+QWL YW+LYSF+TL EL +LEW+P W A
Sbjct: 14 LAGPVVMLLYPLYASVIAIESPSKVDDEQWLAYWILYSFLTLSELILQSLLEWIPIWYTA 73
Query: 675 KLFFNCWLVLPQFNGAAHVYEHFVR 601
KL F WLVLPQF GAA +Y VR
Sbjct: 74 KLVFVAWLVLPQFRGAAFIYNKVVR 98
>sptr|Q07764|Q07764 Abscisic acid and stress inducible (A22) gene.
Length = 130
Score = 115 bits (289), Expect = 9e-25
Identities = 51/95 (53%), Positives = 67/95 (70%)
Frame = -3
Query: 876 LVNNMDVLAGPLVSLAYPLYASVRAIETKSAVDDQQWLTYWVLYSFITLFELTFAPVLEW 697
L+ ++ +AGP ++L YPLYASV A+E+ S VDD+QWL YW+LYSFITL E+ PVL W
Sbjct: 7 LLTHLHSVAGPSITLLYPLYASVCAMESPSKVDDEQWLAYWILYSFITLLEMVAEPVLYW 66
Query: 696 LPFWSYAKLFFNCWLVLPQFNGAAHVYEHFVRPMI 592
+P W KL F WL LPQF GA+ +Y+ VR +
Sbjct: 67 IPVWYPVKLLFVAWLALPQFKGASFIYDKVVREQL 101
>sptrnew|BAC80265|BAC80265 Hypothetical protein Wrab15.
Length = 130
Score = 115 bits (288), Expect = 1e-24
Identities = 51/95 (53%), Positives = 67/95 (70%)
Frame = -3
Query: 876 LVNNMDVLAGPLVSLAYPLYASVRAIETKSAVDDQQWLTYWVLYSFITLFELTFAPVLEW 697
L+ ++ +AGP ++L YPLYASV A+E+ S VDD+QWL YW+LYSFITL E+ PVL W
Sbjct: 7 LLTHLHSVAGPSITLLYPLYASVCAMESPSKVDDEQWLAYWILYSFITLMEMLAEPVLYW 66
Query: 696 LPFWSYAKLFFNCWLVLPQFNGAAHVYEHFVRPMI 592
+P W KL F WL LPQF GA+ +Y+ VR +
Sbjct: 67 IPVWYPVKLLFVAWLALPQFKGASFIYDKVVREQL 101
>sptr|Q9AX25|Q9AX25 P0456A01.10 protein (P0042A10.34 protein).
Length = 128
Score = 114 bits (285), Expect = 3e-24
Identities = 57/118 (48%), Positives = 77/118 (65%), Gaps = 2/118 (1%)
Frame = -3
Query: 810 VRAIETKSAVDDQQWLTYWVLYSFITLFELTFAPVLEWLPFWSYAKLFFNCWLVLPQFNG 631
+RAIE+ S +DDQQWLTYWVLYS ITLFEL+ VL+W P W Y KL F CWLVLP FNG
Sbjct: 1 MRAIESPSTLDDQQWLTYWVLYSLITLFELSCWKVLQWFPLWPYMKLLFCCWLVLPIFNG 60
Query: 630 AAHVYEHFVRPMIVNQQVVNIWYIPKKDESSR--PDDVISAAQRYIEQNGSKAFENLV 463
AA++YE VR Q V+ Y ++ ++ + D + +R+IE +G A + ++
Sbjct: 61 AAYIYETHVRRYFKIGQYVSPNYNERQRKALQMMSLDARKSVERFIESHGPDALDKII 118
>sptr|Q9S760|Q9S760 ATHVA22D (Putative abscisic acid-induced
protein).
Length = 135
Score = 110 bits (276), Expect = 3e-23
Identities = 48/84 (57%), Positives = 61/84 (72%)
Frame = -3
Query: 852 AGPLVSLAYPLYASVRAIETKSAVDDQQWLTYWVLYSFITLFELTFAPVLEWLPFWSYAK 673
AGP+V L YPLYASV A+E+ + VDD+QWL YW++YSF++L EL ++EW+P W K
Sbjct: 15 AGPIVMLLYPLYASVIAMESTTKVDDEQWLAYWIIYSFLSLTELILQSLIEWIPIWYTVK 74
Query: 672 LFFNCWLVLPQFNGAAHVYEHFVR 601
L F WLVLPQF GAA +Y VR
Sbjct: 75 LVFVAWLVLPQFQGAAFIYNRVVR 98
>sptr|Q9LUE9|Q9LUE9 Genomic DNA, chromosome 5, P1 clone:MFB16.
Length = 107
Score = 108 bits (271), Expect = 1e-22
Identities = 50/78 (64%), Positives = 57/78 (73%)
Frame = -3
Query: 834 LAYPLYASVRAIETKSAVDDQQWLTYWVLYSFITLFELTFAPVLEWLPFWSYAKLFFNCW 655
L YPLYASV AIE+ S VDD+QWL YW+LYSF+TL EL +LEW+P W AKL F W
Sbjct: 2 LLYPLYASVIAIESPSKVDDEQWLAYWILYSFLTLSELILQSLLEWIPIWYTAKLVFVAW 61
Query: 654 LVLPQFNGAAHVYEHFVR 601
LVLPQF GAA +Y VR
Sbjct: 62 LVLPQFRGAAFIYNKVVR 79
>sptr|Q9SW30|Q9SW30 Abscisic acid-induced-like protein.
Length = 116
Score = 102 bits (254), Expect = 1e-20
Identities = 44/78 (56%), Positives = 56/78 (71%)
Frame = -3
Query: 834 LAYPLYASVRAIETKSAVDDQQWLTYWVLYSFITLFELTFAPVLEWLPFWSYAKLFFNCW 655
L YPLYASV A+E+ + VDD+QWL YW++YSF++L EL ++EW+P W KL F W
Sbjct: 2 LLYPLYASVIAMESTTKVDDEQWLAYWIIYSFLSLTELILQSLIEWIPIWYTVKLVFVAW 61
Query: 654 LVLPQFNGAAHVYEHFVR 601
LVLPQF GAA +Y VR
Sbjct: 62 LVLPQFQGAAFIYNRVVR 79
>sptr|Q9UU91|Q9UU91 Putative transport protein.
Length = 182
Score = 79.0 bits (193), Expect = 1e-13
Identities = 35/84 (41%), Positives = 52/84 (61%)
Frame = -3
Query: 843 LVSLAYPLYASVRAIETKSAVDDQQWLTYWVLYSFITLFELTFAPVLEWLPFWSYAKLFF 664
L++ A P + S+ AIET + DD QWLTY+++ SF+ + E +L ++P + K F
Sbjct: 62 LLAFAMPAFFSINAIETTNKADDTQWLTYYLVTSFLNVIEYWSQLILYYVPVYWLLKAIF 121
Query: 663 NCWLVLPQFNGAAHVYEHFVRPMI 592
WL LP+FNGA +Y H +RP I
Sbjct: 122 LIWLALPKFNGATIIYRHLIRPYI 145
>sptr|O77388|O77388 Hypothetical 26.0 kDa protein.
Length = 221
Score = 78.2 bits (191), Expect = 2e-13
Identities = 33/88 (37%), Positives = 53/88 (60%)
Frame = -3
Query: 843 LVSLAYPLYASVRAIETKSAVDDQQWLTYWVLYSFITLFELTFAPVLEWLPFWSYAKLFF 664
+V AYP Y S +A+E++S + + WLTYWV++S E +L W+PF+ KL F
Sbjct: 95 VVGFAYPAYQSFKAVESQSRDETKLWLTYWVVFSLFFFIEYLIDIILFWVPFYYLIKLLF 154
Query: 663 NCWLVLPQFNGAAHVYEHFVRPMIVNQQ 580
+L +PQ GA VY + +RP+++ +
Sbjct: 155 LLYLYMPQVRGAVMVYNYIIRPILLKHE 182
>sw|Q60870|DP1_MOUSE Polyposis locus protein 1 homolog (TB2 protein
homolog) (GP106).
Length = 185
Score = 76.6 bits (187), Expect = 6e-13
Identities = 46/135 (34%), Positives = 67/135 (49%), Gaps = 2/135 (1%)
Frame = -3
Query: 843 LVSLAYPLYASVRAIETKSAVDDQQWLTYWVLYSFITLFELTFAPVLEWLPFWSYAKLFF 664
L+ YP Y S++AIE+ + DD QWLTYWV+Y ++ E L WLPF+ K F
Sbjct: 57 LIGFGYPAYISMKAIESPNKDDDTQWLTYWVVYGVFSIAEFFSDLFLSWLPFYYMLKCGF 116
Query: 663 NCWLVLPQ-FNGAAHVYEHFVRPMIV-NQQVVNIWYIPKKDESSRPDDVISAAQRYIEQN 490
W + P NGA +Y +RP+ + ++ V+ KD++ D IS +
Sbjct: 117 LLWCMAPSPANGAEMLYRRIIRPIFLRHESQVDSVVKDVKDKAKETADAIS-------KE 169
Query: 489 GSKAFENLVNKFKSS 445
KA NL+ K S
Sbjct: 170 VKKATVNLLGDVKKS 184
>sw|Q00765|DP1_HUMAN Polyposis locus protein 1 (TB2 protein).
Length = 185
Score = 74.7 bits (182), Expect = 2e-12
Identities = 43/135 (31%), Positives = 67/135 (49%), Gaps = 2/135 (1%)
Frame = -3
Query: 843 LVSLAYPLYASVRAIETKSAVDDQQWLTYWVLYSFITLFELTFAPVLEWLPFWSYAKLFF 664
L+ YP Y S++AIE+ + DD QWLTYWV+Y ++ E L W PF+ K F
Sbjct: 57 LIGFGYPAYISIKAIESPNKEDDTQWLTYWVVYGVFSIAEFFSDIFLSWFPFYYMLKCGF 116
Query: 663 NCWLVLPQ-FNGAAHVYEHFVRPMIV-NQQVVNIWYIPKKDESSRPDDVISAAQRYIEQN 490
W + P NGA +Y+ +RP + ++ ++ KD++ D I+ +
Sbjct: 117 LLWCMAPSPSNGAELLYKRIIRPFFLKHESQMDSVVKDLKDKAKETADAIT-------KE 169
Query: 489 GSKAFENLVNKFKSS 445
KA NL+ + K S
Sbjct: 170 AKKATVNLLGEEKKS 184
>sptr|Q9CQG4|Q9CQG4 Deleted in polyposis 1.
Length = 185
Score = 74.3 bits (181), Expect = 3e-12
Identities = 45/135 (33%), Positives = 66/135 (48%), Gaps = 2/135 (1%)
Frame = -3
Query: 843 LVSLAYPLYASVRAIETKSAVDDQQWLTYWVLYSFITLFELTFAPVLEWLPFWSYAKLFF 664
L+ YP Y S++AIE+ + DD QWLTYWV+Y ++ E L W PF+ K F
Sbjct: 57 LIGFGYPAYISMKAIESPNKDDDTQWLTYWVVYGVFSIAEFFSDLFLSWFPFYYMLKCGF 116
Query: 663 NCWLVLPQ-FNGAAHVYEHFVRPMIV-NQQVVNIWYIPKKDESSRPDDVISAAQRYIEQN 490
W + P NGA +Y +RP+ + ++ V+ KD++ D IS +
Sbjct: 117 LLWCMAPSPANGAEMLYRRIIRPIFLKHESQVDSVVKDVKDKAKETADAIS-------KE 169
Query: 489 GSKAFENLVNKFKSS 445
KA NL+ K S
Sbjct: 170 VKKATVNLLGDEKKS 184
>sptr|Q96LM0|Q96LM0 Hypothetical protein FLJ25383 (TB2 protein-like
1).
Length = 184
Score = 73.6 bits (179), Expect = 5e-12
Identities = 35/86 (40%), Positives = 51/86 (59%), Gaps = 1/86 (1%)
Frame = -3
Query: 843 LVSLAYPLYASVRAIETKSAVDDQQWLTYWVLYSFITLFELTFAPVLEWLPFWSYAKLFF 664
L+ YP YAS++AIE+ S DD WLTYWV+Y+ L E +L W PF+ K F
Sbjct: 60 LIGFVYPAYASIKAIESPSKDDDTVWLTYWVVYALFGLAEFFSDLLLSWFPFYYVGKCAF 119
Query: 663 NCWLVLPQ-FNGAAHVYEHFVRPMIV 589
+ + P+ +NGA +Y+ VRP+ +
Sbjct: 120 LLFCMAPRPWNGALMLYQRVVRPLFL 145
>sptr|Q96HR9|Q96HR9 Hypothetical protein.
Length = 184
Score = 73.6 bits (179), Expect = 5e-12
Identities = 35/86 (40%), Positives = 51/86 (59%), Gaps = 1/86 (1%)
Frame = -3
Query: 843 LVSLAYPLYASVRAIETKSAVDDQQWLTYWVLYSFITLFELTFAPVLEWLPFWSYAKLFF 664
L+ YP YAS++AIE+ S DD WLTYWV+Y+ L E +L W PF+ K F
Sbjct: 60 LIGFVYPAYASIKAIESPSKDDDTVWLTYWVVYALFGLAEFFSDLLLSWFPFYYVGKCAF 119
Query: 663 NCWLVLPQ-FNGAAHVYEHFVRPMIV 589
+ + P+ +NGA +Y+ VRP+ +
Sbjct: 120 LLFCMAPRPWNGALMLYQRVVRPLFL 145
>sptr|Q921E4|Q921E4 Deleted in polyposis 1.
Length = 185
Score = 72.8 bits (177), Expect = 9e-12
Identities = 45/135 (33%), Positives = 65/135 (48%), Gaps = 2/135 (1%)
Frame = -3
Query: 843 LVSLAYPLYASVRAIETKSAVDDQQWLTYWVLYSFITLFELTFAPVLEWLPFWSYAKLFF 664
L+ YP Y S++AIE+ + DD QWLTYWV+Y ++ E L W PF+ K F
Sbjct: 57 LIGFGYPAYISMKAIESPNKDDDTQWLTYWVVYGVFSIAEFFSDLFLSWFPFYYMLKCGF 116
Query: 663 NCWLVLPQ-FNGAAHVYEHFVRPMIV-NQQVVNIWYIPKKDESSRPDDVISAAQRYIEQN 490
W + P NGA Y +RP+ + ++ V+ KD++ D IS +
Sbjct: 117 LLWCMAPSPANGAEMRYRRIIRPIFLKHESQVDSVVKDVKDKAKETADAIS-------KE 169
Query: 489 GSKAFENLVNKFKSS 445
KA NL+ K S
Sbjct: 170 VKKATVNLLGDEKKS 184
>sptr|O45108|O45108 Hypothetical 22.8 kDa protein.
Length = 205
Score = 72.4 bits (176), Expect = 1e-11
Identities = 32/85 (37%), Positives = 46/85 (54%)
Frame = -3
Query: 843 LVSLAYPLYASVRAIETKSAVDDQQWLTYWVLYSFITLFELTFAPVLEWLPFWSYAKLFF 664
L+ YP YASV+AI + DD WL YW ++ + L + +L W PF+ AK F
Sbjct: 106 LIGFGYPTYASVKAIRSPGGDDDTVWLIYWTCFAVLYLVDFFSEAILSWFPFYYIAKACF 165
Query: 663 NCWLVLPQFNGAAHVYEHFVRPMIV 589
+L LPQ G+ YE V P+++
Sbjct: 166 LVYLYLPQTQGSVMFYETIVDPLVI 190
>sptrnew|EAA31781|EAA31781 Hypothetical protein.
Length = 168
Score = 72.4 bits (176), Expect = 1e-11
Identities = 37/99 (37%), Positives = 51/99 (51%)
Frame = -3
Query: 888 FLKVLVNNMDVLAGPLVSLAYPLYASVRAIETKSAVDDQQWLTYWVLYSFITLFELTFAP 709
F ++ N L L P Y S+ A+ T S DD QWLTYWV++S T+ E +
Sbjct: 48 FFLIIFNLGGQLLTNLAGFVLPGYYSLNALFTASKQDDTQWLTYWVVFSLFTVIE-SLIS 106
Query: 708 VLEWLPFWSYAKLFFNCWLVLPQFNGAAHVYEHFVRPMI 592
V+ W PF+ K F WL LP F GA ++ F+ P +
Sbjct: 107 VVYWFPFYFTFKFVFLLWLSLPTFKGAETIFRSFLAPTL 145
>sptr|Q871R7|Q871R7 Related to Ypt-interacting protein YIP2.
Length = 168
Score = 72.4 bits (176), Expect = 1e-11
Identities = 37/99 (37%), Positives = 51/99 (51%)
Frame = -3
Query: 888 FLKVLVNNMDVLAGPLVSLAYPLYASVRAIETKSAVDDQQWLTYWVLYSFITLFELTFAP 709
F ++ N L L P Y S+ A+ T S DD QWLTYWV++S T+ E +
Sbjct: 48 FFLIIFNLGGQLLTNLAGFVLPGYYSLNALFTASKQDDTQWLTYWVVFSLFTVIE-SLIS 106
Query: 708 VLEWLPFWSYAKLFFNCWLVLPQFNGAAHVYEHFVRPMI 592
V+ W PF+ K F WL LP F GA ++ F+ P +
Sbjct: 107 VVYWFPFYFTFKFVFLLWLSLPTFKGAETIFRSFLAPTL 145
>sptrnew|AAH06218|AAH06218 Hypothetical protein.
Length = 252
Score = 72.0 bits (175), Expect = 1e-11
Identities = 32/97 (32%), Positives = 55/97 (56%)
Frame = -3
Query: 828 YPLYASVRAIETKSAVDDQQWLTYWVLYSFITLFELTFAPVLEWLPFWSYAKLFFNCWLV 649
YP Y+S +A++TK+ + +W+ YW++++F T E VL W PF+ K+ F WL+
Sbjct: 18 YPAYSSYKAVKTKNVKEYVKWMMYWIVFAFFTTAETLTDIVLSWFPFYFELKIAFVIWLL 77
Query: 648 LPQFNGAAHVYEHFVRPMIVNQQVVNIWYIPKKDESS 538
P G++ +Y FV P + N++ YI + + S
Sbjct: 78 SPYTKGSSVLYRKFVHPTLSNKEKEIDEYITQARDKS 114
>sptr|Q9N4G8|Q9N4G8 Hypothetical 20.6 kDa protein.
Length = 183
Score = 72.0 bits (175), Expect = 1e-11
Identities = 31/82 (37%), Positives = 49/82 (59%)
Frame = -3
Query: 840 VSLAYPLYASVRAIETKSAVDDQQWLTYWVLYSFITLFELTFAPVLEWLPFWSYAKLFFN 661
+ YP Y S++AIE+ + DD QWLTYWV+++ +++ E ++ P + K F
Sbjct: 67 MGFVYPAYMSIKAIESSNKEDDTQWLTYWVIFAILSVVEFFSVQIVAVFPVYWLFKSIFL 126
Query: 660 CWLVLPQFNGAAHVYEHFVRPM 595
+L LP F GAA +Y FV+P+
Sbjct: 127 LYLYLPSFLGAAKLYHRFVKPV 148
>sptrnew|EAA05239|EAA05239 ENSANGP00000018512 (Fragment).
Length = 321
Score = 71.2 bits (173), Expect = 3e-11
Identities = 29/88 (32%), Positives = 49/88 (55%)
Frame = -3
Query: 843 LVSLAYPLYASVRAIETKSAVDDQQWLTYWVLYSFITLFELTFAPVLEWLPFWSYAKLFF 664
L YP YAS +A+ TK+ + +W+ YW++++F T E +L W PF+ K+
Sbjct: 77 LFGTLYPAYASYKAVRTKNVKEYVKWMMYWIVFAFFTFIETFTDILLSWFPFYYEIKVIV 136
Query: 663 NCWLVLPQFNGAAHVYEHFVRPMIVNQQ 580
WL+ P G++ +Y FV PM+ ++
Sbjct: 137 VLWLLSPATRGSSTLYRKFVHPMLTRRE 164
>sptr|Q9JM62|Q9JM62 Polyposis locus protein 1-like 1 (TB2
protein-like 1).
Length = 201
Score = 70.1 bits (170), Expect = 6e-11
Identities = 34/90 (37%), Positives = 50/90 (55%), Gaps = 1/90 (1%)
Frame = -3
Query: 843 LVSLAYPLYASVRAIETKSAVDDQQWLTYWVLYSFITLFELTFAPVLEWLPFWSYAKLFF 664
++ YP YASV+AIE+ S DD WLTYWV+Y+ L E +L W PF+ K F
Sbjct: 60 VIGFVYPAYASVKAIESPSKEDDTVWLTYWVVYALFGLVEFFSDLLLFWFPFYYAGKCAF 119
Query: 663 NCWLVLP-QFNGAAHVYEHFVRPMIVNQQV 577
+ + P +NGA +Y +RP+ + +
Sbjct: 120 LLFCMTPGPWNGALLLYHRVIRPLFLKHHM 149
>sptr|Q8K0X2|Q8K0X2 Hypothetical protein.
Length = 174
Score = 70.1 bits (170), Expect = 6e-11
Identities = 34/90 (37%), Positives = 50/90 (55%), Gaps = 1/90 (1%)
Frame = -3
Query: 843 LVSLAYPLYASVRAIETKSAVDDQQWLTYWVLYSFITLFELTFAPVLEWLPFWSYAKLFF 664
++ YP YASV+AIE+ S DD WLTYWV+Y+ L E +L W PF+ K F
Sbjct: 60 VIGFVYPAYASVKAIESPSKEDDTVWLTYWVVYALFGLVEFFSDLLLFWFPFYYAGKCAF 119
Query: 663 NCWLVLP-QFNGAAHVYEHFVRPMIVNQQV 577
+ + P +NGA +Y +RP+ + +
Sbjct: 120 LLFCMTPGPWNGALLLYHRVIRPLFLKHHM 149
>sptr|Q8MLS9|Q8MLS9 CG30193-PA (RE29641p).
Length = 288
Score = 69.3 bits (168), Expect = 1e-10
Identities = 28/83 (33%), Positives = 46/83 (55%)
Frame = -3
Query: 828 YPLYASVRAIETKSAVDDQQWLTYWVLYSFITLFELTFAPVLEWLPFWSYAKLFFNCWLV 649
YP YAS +A+ TK + +W+ YW++++F T E + WLPF+ K+ WL+
Sbjct: 18 YPAYASYKAVRTKDVKEYVKWMMYWIVFAFFTCIETFTDIFISWLPFYYEVKVALVFWLL 77
Query: 648 LPQFNGAAHVYEHFVRPMIVNQQ 580
P G++ +Y FV PM+ +
Sbjct: 78 SPATKGSSTLYRKFVHPMLTRHE 100
>sptr|Q8SX60|Q8SX60 LD42159p (CG30193-PD).
Length = 435
Score = 69.3 bits (168), Expect = 1e-10
Identities = 28/83 (33%), Positives = 46/83 (55%)
Frame = -3
Query: 828 YPLYASVRAIETKSAVDDQQWLTYWVLYSFITLFELTFAPVLEWLPFWSYAKLFFNCWLV 649
YP YAS +A+ TK + +W+ YW++++F T E + WLPF+ K+ WL+
Sbjct: 165 YPAYASYKAVRTKDVKEYVKWMMYWIVFAFFTCIETFTDIFISWLPFYYEVKVALVFWLL 224
Query: 648 LPQFNGAAHVYEHFVRPMIVNQQ 580
P G++ +Y FV PM+ +
Sbjct: 225 SPATKGSSTLYRKFVHPMLTRHE 247
>sptrnew|EAA13321|EAA13321 AgCP2771 (Fragment).
Length = 164
Score = 68.6 bits (166), Expect = 2e-10
Identities = 29/83 (34%), Positives = 54/83 (65%), Gaps = 1/83 (1%)
Frame = -3
Query: 843 LVSLAYPLYASVRAIETKSAVDDQQWLTYWVLYSFITLFELTFAPVLEWLPFWSYAKLFF 664
++ +AYP Y S++AIET++ DD +WLTYWV + +++FE +++ +PF+ K F
Sbjct: 48 VIGVAYPAYISMKAIETRTKEDDTKWLTYWVTFGVLSVFEHFSFFLVQIIPFYWLLKCLF 107
Query: 663 NCWLVLP-QFNGAAHVYEHFVRP 598
+ W ++P + NG+ +Y ++P
Sbjct: 108 HIWCMVPMENNGSTIMYHKVIQP 130
Database: /db/trembl-ebi/tmp/swall
Posted date: Sep 26, 2003 8:29 PM
Number of letters in database: 406,886,216
Number of sequences in database: 1,272,877
Lambda K H
0.318 0.135 0.401
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 795,622,325
Number of Sequences: 1272877
Number of extensions: 16990496
Number of successful extensions: 43871
Number of sequences better than 10.0: 30
Number of HSP's better than 10.0 without gapping: 41751
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 43846
length of database: 406,886,216
effective HSP length: 124
effective length of database: 249,049,468
effective search space used: 63507614340
frameshift window, decay const: 50, 0.1
T: 12
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)

