BLASTX 2.2.6 [Apr-09-2003]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= 131537.2.723
(1314 letters)
Database: /db/trembl-ebi/tmp/swall
1,288,562 sequences; 411,996,502 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
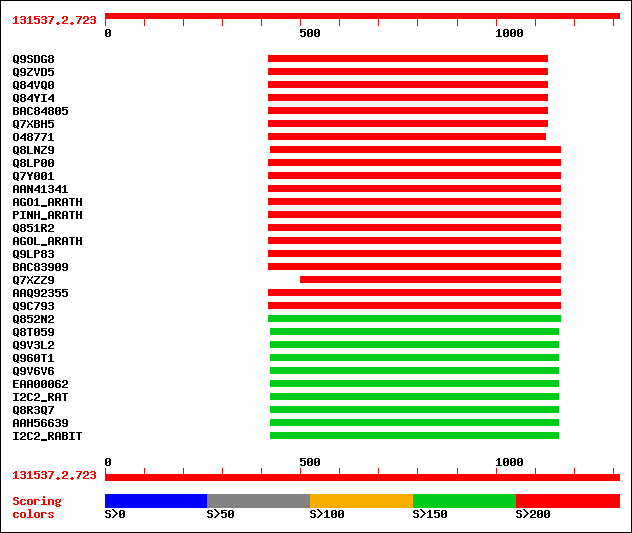
sptr|Q9SDG8|Q9SDG8 ESTs AU068544(C30430). 393 e-108
sptr|Q9ZVD5|Q9ZVD5 Argonaute (AGO1)-like protein (Putative post-... 383 e-105
sptr|Q84VQ0|Q84VQ0 PAZ (Piwi Argonaut and Zwille) family. 378 e-103
sptr|Q84YI4|Q84YI4 ARGONAUTE9 protein. 376 e-103
sptrnew|BAC84805|BAC84805 Putative ARGONAUTE9 protein. 320 4e-86
sptr|Q7XBH5|Q7XBH5 Zwille pinhead-like protein. 318 1e-85
sptr|O48771|O48771 Argonaute (AGO1)-like protein. 313 4e-84
sptr|Q8LNZ9|Q8LNZ9 AGO1 homologous protein. 242 8e-63
sptr|Q8LP00|Q8LP00 ZLL/PNH homologous protein. 241 2e-62
sptr|Q7Y001|Q7Y001 Putative leaf development and shoot apical me... 239 7e-62
sptrnew|AAN41341|AAN41341 Putative leaf development protein Argo... 238 1e-61
sw|O04379|AGO1_ARATH Argonaute protein. 238 1e-61
sw|Q9XGW1|PINH_ARATH PINHEAD protein (ZWILLE protein). 235 9e-61
sptr|Q851R2|Q851R2 Putative argonaute protein. 233 4e-60
sw|Q9SJK3|AGOL_ARATH Argonaute-like protein At2g27880. 227 3e-58
sptr|Q9LP83|Q9LP83 T1N15.2. 224 2e-57
sptrnew|BAC83909|BAC83909 Putative leaf development protein Argo... 216 5e-55
sptr|Q7XZZ9|Q7XZZ9 Putative leaf development and shoot apical me... 207 2e-52
sptrnew|AAQ92355|AAQ92355 ZIPPY. 201 3e-50
sptr|Q9C793|Q9C793 Pinhead-like protein. 201 3e-50
sptr|Q852N2|Q852N2 Putative argonaute protein. 195 1e-48
sptr|Q8T059|Q8T059 LD26301p. 191 2e-47
sptr|Q9V3L2|Q9V3L2 AGO1 protein. 190 5e-47
sptr|Q960T1|Q960T1 LD36719p. 190 5e-47
sptr|Q9V6V6|Q9V6V6 AGO1 protein. 190 5e-47
sptrnew|EAA00062|EAA00062 EbiP8896 (Fragment). 189 8e-47
sw|Q9QZ81|I2C2_RAT Eukaryotic translation initiation factor 2C 2... 189 1e-46
sptr|Q8R3Q7|Q8R3Q7 Similar to eukaryotic translation initiation ... 188 2e-46
sptrnew|AAH56639|AAH56639 Hypothetical protein (Fragment). 188 2e-46
sw|O77503|I2C2_RABIT Eukaryotic translation initiation factor 2C... 188 2e-46
>sptr|Q9SDG8|Q9SDG8 ESTs AU068544(C30430).
Length = 904
Score = 393 bits (1010), Expect = e-108
Identities = 190/240 (79%), Positives = 216/240 (90%), Gaps = 2/240 (0%)
Frame = +2
Query: 419 YLLNLLMKINAKLGGLNSLLQVEASSSIPHVSQVPTIILGMDVSHGHPGQ-DRPSVAAVV 595
Y+ N+L+KINAKLGGLNSLLQ+E S SIP VS+VPTIILGMDVSHG PGQ D PS+AAVV
Sbjct: 598 YITNVLLKINAKLGGLNSLLQIETSPSIPLVSKVPTIILGMDVSHGSPGQSDIPSIAAVV 657
Query: 596 SSRQWPLISRYRASVHTQSARLEMMSSLFKPRGT-DDDGLIRESLIDFYTSSGKRKPEHI 772
SSR+WPL+S+YRASV +QS +LEM+ LFKP+G +DDGLIRE L+DFYTS+GKRKP+ +
Sbjct: 658 SSREWPLVSKYRASVRSQSPKLEMIDGLFKPQGAQEDDGLIRELLVDFYTSTGKRKPDQV 717
Query: 773 IIFRDGVSESQFTQVINIELDQIIEACKFLDEKWSPKFTVIVAQKNHHTKFFQTASPDNV 952
IIFRDGVSESQFTQV+NIELDQIIEACKFLDE WSPKFT+IVAQKNHHTKFF S +NV
Sbjct: 718 IIFRDGVSESQFTQVLNIELDQIIEACKFLDENWSPKFTLIVAQKNHHTKFFVPGSQNNV 777
Query: 953 LPGTVVDSKVCHPKNFDFYMCAHAGMIGTTRPTHYHVLHDEIGFSADEMQEFVHSLSYVY 1132
PGTVVD+ VCHP+N DFYMCAHAGMIGTTRPTHYH+LHDEIGFSAD++QE VHSLSYVY
Sbjct: 778 PPGTVVDNAVCHPRNNDFYMCAHAGMIGTTRPTHYHILHDEIGFSADDLQELVHSLSYVY 837
>sptr|Q9ZVD5|Q9ZVD5 Argonaute (AGO1)-like protein (Putative
post-transcriptional gene silencing protein).
Length = 924
Score = 383 bits (984), Expect = e-105
Identities = 182/239 (76%), Positives = 210/239 (87%), Gaps = 1/239 (0%)
Frame = +2
Query: 419 YLLNLLMKINAKLGGLNSLLQVEASSSIPHVSQVPTIILGMDVSHGHPGQ-DRPSVAAVV 595
YL NLL+KINAKLGGLNS+L VE + + +S+VPTIILGMDVSHG PGQ D PS+AAVV
Sbjct: 619 YLTNLLLKINAKLGGLNSMLSVERTPAFTVISKVPTIILGMDVSHGSPGQSDVPSIAAVV 678
Query: 596 SSRQWPLISRYRASVHTQSARLEMMSSLFKPRGTDDDGLIRESLIDFYTSSGKRKPEHII 775
SSR+WPLIS+YRASV TQ ++ EM+ SL K GT+DDG+I+E L+DFYTSS KRKPEHII
Sbjct: 679 SSREWPLISKYRASVRTQPSKAEMIESLVKKNGTEDDGIIKELLVDFYTSSNKRKPEHII 738
Query: 776 IFRDGVSESQFTQVINIELDQIIEACKFLDEKWSPKFTVIVAQKNHHTKFFQTASPDNVL 955
IFRDGVSESQF QV+NIELDQIIEACK LD W+PKF ++VAQKNHHTKFFQ SP+NV
Sbjct: 739 IFRDGVSESQFNQVLNIELDQIIEACKLLDANWNPKFLLLVAQKNHHTKFFQPTSPENVP 798
Query: 956 PGTVVDSKVCHPKNFDFYMCAHAGMIGTTRPTHYHVLHDEIGFSADEMQEFVHSLSYVY 1132
PGT++D+K+CHPKN DFY+CAHAGMIGTTRPTHYHVL+DEIGFSADE+QE VHSLSYVY
Sbjct: 799 PGTIIDNKICHPKNNDFYLCAHAGMIGTTRPTHYHVLYDEIGFSADELQELVHSLSYVY 857
>sptr|Q84VQ0|Q84VQ0 PAZ (Piwi Argonaut and Zwille) family.
Length = 892
Score = 378 bits (970), Expect = e-103
Identities = 177/239 (74%), Positives = 210/239 (87%), Gaps = 1/239 (0%)
Frame = +2
Query: 419 YLLNLLMKINAKLGGLNSLLQVEASSSIPHVSQVPTIILGMDVSHGHPGQ-DRPSVAAVV 595
YL N+L+KINAKLGGLNSLL +E S ++P V+QVPTII+GMDVSHG PGQ D PS+AAVV
Sbjct: 587 YLTNVLLKINAKLGGLNSLLAMERSPAMPKVTQVPTIIVGMDVSHGSPGQSDIPSIAAVV 646
Query: 596 SSRQWPLISRYRASVHTQSARLEMMSSLFKPRGTDDDGLIRESLIDFYTSSGKRKPEHII 775
SSRQWPLIS+Y+A V TQS ++EM+ +LFKP D+G+ RE L+DFY SS RKPEHII
Sbjct: 647 SSRQWPLISKYKACVRTQSRKMEMIDNLFKPVNGKDEGMFRELLLDFYYSSENRKPEHII 706
Query: 776 IFRDGVSESQFTQVINIELDQIIEACKFLDEKWSPKFTVIVAQKNHHTKFFQTASPDNVL 955
IFRDGVSESQF QV+NIELDQ+++ACKFLD+ W PKFTVIVAQKNHHTKFFQ+ PDNV
Sbjct: 707 IFRDGVSESQFNQVLNIELDQMMQACKFLDDTWHPKFTVIVAQKNHHTKFFQSRGPDNVP 766
Query: 956 PGTVVDSKVCHPKNFDFYMCAHAGMIGTTRPTHYHVLHDEIGFSADEMQEFVHSLSYVY 1132
PGT++DS++CHP+NFDFY+CAHAGMIGTTRPTHYHVL+DEIGF+ D++QE VHSLSYVY
Sbjct: 767 PGTIIDSQICHPRNFDFYLCAHAGMIGTTRPTHYHVLYDEIGFATDDLQELVHSLSYVY 825
>sptr|Q84YI4|Q84YI4 ARGONAUTE9 protein.
Length = 896
Score = 376 bits (965), Expect = e-103
Identities = 176/239 (73%), Positives = 210/239 (87%), Gaps = 1/239 (0%)
Frame = +2
Query: 419 YLLNLLMKINAKLGGLNSLLQVEASSSIPHVSQVPTIILGMDVSHGHPGQ-DRPSVAAVV 595
YL N+L+KINAKLGGLNSLL +E S ++P V+QVPTII+GMDVSHG PGQ D PS+AAVV
Sbjct: 591 YLTNVLLKINAKLGGLNSLLAMERSPAMPKVTQVPTIIVGMDVSHGSPGQSDIPSIAAVV 650
Query: 596 SSRQWPLISRYRASVHTQSARLEMMSSLFKPRGTDDDGLIRESLIDFYTSSGKRKPEHII 775
SSRQWPLIS+Y+A V TQS ++EM+ +LFKP D+G+ RE L+DFY SS RKPEHII
Sbjct: 651 SSRQWPLISKYKACVRTQSRKMEMIDNLFKPVNGKDEGMFRELLLDFYYSSENRKPEHII 710
Query: 776 IFRDGVSESQFTQVINIELDQIIEACKFLDEKWSPKFTVIVAQKNHHTKFFQTASPDNVL 955
IFRDGVSESQF QV+NIELDQ+++ACKFLD+ W PKFTVIVAQKNHHTKFFQ+ PDNV
Sbjct: 711 IFRDGVSESQFNQVLNIELDQMMQACKFLDDTWHPKFTVIVAQKNHHTKFFQSRGPDNVP 770
Query: 956 PGTVVDSKVCHPKNFDFYMCAHAGMIGTTRPTHYHVLHDEIGFSADEMQEFVHSLSYVY 1132
PGT++DS++CHP+NFDFY+CAHAGMIGTTRPTHYHVL+DEIGF+ D++QE VHSLS+VY
Sbjct: 771 PGTIIDSQICHPRNFDFYLCAHAGMIGTTRPTHYHVLYDEIGFATDDLQELVHSLSHVY 829
>sptrnew|BAC84805|BAC84805 Putative ARGONAUTE9 protein.
Length = 889
Score = 320 bits (819), Expect = 4e-86
Identities = 161/240 (67%), Positives = 195/240 (81%), Gaps = 2/240 (0%)
Frame = +2
Query: 419 YLLNLLMKINAKLGGLNSLLQVEASSSIPHVSQVPTIILGMDVSHGHPGQ-DRPSVAAVV 595
Y N+L+KINAKLGG+NS L +E IP V+Q PT+ILGMDVSHG PG+ D PS+AAVV
Sbjct: 583 YYTNVLLKINAKLGGMNSKLSLEHRHMIPIVNQTPTLILGMDVSHGSPGRADVPSIAAVV 642
Query: 596 SSRQWPLISRYRASVHTQSARLEMMSSLFKPRGTD-DDGLIRESLIDFYTSSGKRKPEHI 772
SR WPLISRYRASV TQS ++EM+ SLFKP DDG+IRE L+DFY +S +RKP+ I
Sbjct: 643 GSRCWPLISRYRASVRTQSPKVEMIDSLFKPLDDGKDDGIIRELLLDFYKTSQQRKPKQI 702
Query: 773 IIFRDGVSESQFTQVINIELDQIIEACKFLDEKWSPKFTVIVAQKNHHTKFFQTASPDNV 952
IIFRDGVSESQF+QV+N+EL+QII+A +++D+ PKFTVI+AQKNHHTK FQ +PDNV
Sbjct: 703 IIFRDGVSESQFSQVLNVELNQIIKAYQYMDQGPIPKFTVIIAQKNHHTKLFQENTPDNV 762
Query: 953 LPGTVVDSKVCHPKNFDFYMCAHAGMIGTTRPTHYHVLHDEIGFSADEMQEFVHSLSYVY 1132
PGTVVDS + HP+ +DFYM AHAG IGT+RPTHYHVL DEIGF D++Q+ V SLSYVY
Sbjct: 763 PPGTVVDSGIVHPRQYDFYMYAHAGPIGTSRPTHYHVLLDEIGFLPDDVQKLVLSLSYVY 822
>sptr|Q7XBH5|Q7XBH5 Zwille pinhead-like protein.
Length = 361
Score = 318 bits (815), Expect = 1e-85
Identities = 160/240 (66%), Positives = 194/240 (80%), Gaps = 2/240 (0%)
Frame = +2
Query: 419 YLLNLLMKINAKLGGLNSLLQVEASSSIPHVSQVPTIILGMDVSHGHPGQ-DRPSVAAVV 595
Y N+L+KINAKLGG+NS L +E IP V+Q PT+ILGMDVSHG PG+ D PS+AAV
Sbjct: 57 YYTNVLLKINAKLGGMNSKLSLEHRHMIPIVNQTPTLILGMDVSHGSPGRADVPSIAAVA 116
Query: 596 SSRQWPLISRYRASVHTQSARLEMMSSLFKPRGTD-DDGLIRESLIDFYTSSGKRKPEHI 772
SR WPLISRYRASV TQS ++EM+ SLFKP DDG+IRE L+DFY +S +RKP+ I
Sbjct: 117 GSRCWPLISRYRASVRTQSPKVEMIDSLFKPLDDGKDDGIIRELLLDFYKTSQQRKPKQI 176
Query: 773 IIFRDGVSESQFTQVINIELDQIIEACKFLDEKWSPKFTVIVAQKNHHTKFFQTASPDNV 952
IIFRDGVSESQF+QV+N+EL+QII+A +++D+ PKFTVI+AQKNHHTK FQ +PDNV
Sbjct: 177 IIFRDGVSESQFSQVLNVELNQIIKAYQYMDQGPIPKFTVIIAQKNHHTKLFQENTPDNV 236
Query: 953 LPGTVVDSKVCHPKNFDFYMCAHAGMIGTTRPTHYHVLHDEIGFSADEMQEFVHSLSYVY 1132
PGTVVDS + HP+ +DFYM AHAG IGT+RPTHYHVL DEIGF D++Q+ V SLSYVY
Sbjct: 237 PPGTVVDSGIVHPRQYDFYMYAHAGPIGTSRPTHYHVLLDEIGFLPDDVQKLVLSLSYVY 296
>sptr|O48771|O48771 Argonaute (AGO1)-like protein.
Length = 887
Score = 313 bits (802), Expect = 4e-84
Identities = 157/240 (65%), Positives = 193/240 (80%), Gaps = 4/240 (1%)
Frame = +2
Query: 419 YLLNLLMKINAKLGGLNSLLQVEASSSIPHVSQVPTIILGMDVSHGHPGQ-DRPSVAAVV 595
YL N+L+KIN+KLGG+NSLL +E S +IP ++++PT+ILGMDVSHG PG+ D PSVAAVV
Sbjct: 583 YLTNVLLKINSKLGGINSLLGIEYSYNIPLINKIPTLILGMDVSHGPPGRADVPSVAAVV 642
Query: 596 SSRQWPLISRYRASVHTQSARLEMMSSLFKP-RGTD--DDGLIRESLIDFYTSSGKRKPE 766
S+ WPLISRYRA+V TQS RLEM+ SLF+P T+ D+G++ E ++FY +S RKP+
Sbjct: 643 GSKCWPLISRYRAAVRTQSPRLEMIDSLFQPIENTEKGDNGIMNELFVEFYRTSRARKPK 702
Query: 767 HIIIFRDGVSESQFTQVINIELDQIIEACKFLDEKWSPKFTVIVAQKNHHTKFFQTASPD 946
IIIFRDGVSESQF QV+ IE+DQII+A + L E PKFTVIVAQKNHHTK FQ P+
Sbjct: 703 QIIIFRDGVSESQFEQVLKIEVDQIIKAYQRLGESDVPKFTVIVAQKNHHTKLFQAKGPE 762
Query: 947 NVLPGTVVDSKVCHPKNFDFYMCAHAGMIGTTRPTHYHVLHDEIGFSADEMQEFVHSLSY 1126
NV GTVVD+K+ HP N+DFYMCAHAG IGT+RP HYHVL DEIGFS D++Q +HSLSY
Sbjct: 763 NVPAGTVVDTKIVHPTNYDFYMCAHAGKIGTSRPAHYHVLLDEIGFSPDDLQNLIHSLSY 822
>sptr|Q8LNZ9|Q8LNZ9 AGO1 homologous protein.
Length = 909
Score = 242 bits (618), Expect = 8e-63
Identities = 128/262 (48%), Positives = 168/262 (64%), Gaps = 14/262 (5%)
Frame = +2
Query: 422 LLNLLMKINAKLGGLNSLLQVEASSSIPHVSQVPTIILGMDVSHGHPGQDR-PSVAAVVS 598
L NL +KIN K+GG N++L S IP V+ PTII G DV+H HPG+D PS+AAVV+
Sbjct: 580 LANLALKINVKVGGRNTVLVDAVSRRIPLVTDRPTIIFGADVTHPHPGEDSSPSIAAVVA 639
Query: 599 SRQWPLISRYRASVHTQSARLEMMSSLFK-----PRGTDDDGLIRESLIDFYTSSGKRKP 763
S+ WP +++Y V Q+ R E++ L+K RGT G+IRE LI F S+G+ KP
Sbjct: 640 SQDWPEVTKYAGLVSAQAHRQELIEDLYKIWQDPQRGTVSGGMIRELLISFKRSTGE-KP 698
Query: 764 EHIIIFRDGVSESQFTQVINIELDQIIEACKFLDEKWSPKFTVIVAQKNHHTKFFQTASP 943
+ II +RDGVSE QF QV+ EL+ I +AC L+ + PK T IV QK HHT+ F
Sbjct: 699 QRIIFYRDGVSEGQFYQVLLYELNAIRKACASLETNYQPKVTFIVVQKRHHTRLFAHNHN 758
Query: 944 D--------NVLPGTVVDSKVCHPKNFDFYMCAHAGMIGTTRPTHYHVLHDEIGFSADEM 1099
D N+LPGTVVDSK+CHP FDFY+C+HAG+ GT+RP HYHVL +E F+AD +
Sbjct: 759 DQNSVDRSGNILPGTVVDSKICHPTEFDFYLCSHAGIKGTSRPAHYHVLWNENNFTADAL 818
Query: 1100 QEFVHSLSYVYTIADRWIEIAP 1165
Q ++L Y Y R + I P
Sbjct: 819 QILTNNLCYTYARCTRSVSIVP 840
>sptr|Q8LP00|Q8LP00 ZLL/PNH homologous protein.
Length = 978
Score = 241 bits (615), Expect = 2e-62
Identities = 127/263 (48%), Positives = 167/263 (63%), Gaps = 14/263 (5%)
Frame = +2
Query: 419 YLLNLLMKINAKLGGLNSLLQVEASSSIPHVSQVPTIILGMDVSHGHPGQDR-PSVAAVV 595
YL N+ +KIN K+GG N++L S IP VS +PTII G DV+H G+D PS+AAVV
Sbjct: 662 YLANVSLKINVKMGGRNTVLLDAISWRIPLVSDIPTIIFGADVTHPETGEDSSPSIAAVV 721
Query: 596 SSRQWPLISRYRASVHTQSARLEMMSSLFKP-----RGTDDDGLIRESLIDFYTSSGKRK 760
+S+ WP +++Y V Q+ R E++ L+K RGT G+IRE LI F ++G+ K
Sbjct: 722 ASQDWPEVTKYAGLVCAQAHRQELIQDLYKTWHDPQRGTVTGGMIRELLISFRKATGQ-K 780
Query: 761 PEHIIIFRDGVSESQFTQVINIELDQIIEACKFLDEKWSPKFTVIVAQKNHHTKFFQTAS 940
P II +RDGVSE QF QV+ ELD I +AC L+ + P T +V QK HHT+ F
Sbjct: 781 PLRIIFYRDGVSEGQFYQVLLYELDAIRKACASLEPIYQPPVTFVVVQKRHHTRLFANNH 840
Query: 941 PD--------NVLPGTVVDSKVCHPKNFDFYMCAHAGMIGTTRPTHYHVLHDEIGFSADE 1096
D N+LPGTVVDSK+CHP FDFY+C+HAG+ GT+RP HYHVL DE F+ADE
Sbjct: 841 KDRSSTDKSGNILPGTVVDSKICHPSEFDFYLCSHAGIQGTSRPAHYHVLWDENNFTADE 900
Query: 1097 MQEFVHSLSYVYTIADRWIEIAP 1165
MQ ++L Y Y R + + P
Sbjct: 901 MQTLTNNLCYTYARCTRSVSVVP 923
>sptr|Q7Y001|Q7Y001 Putative leaf development and shoot apical
meristem regulating protein.
Length = 1055
Score = 239 bits (610), Expect = 7e-62
Identities = 122/261 (46%), Positives = 168/261 (64%), Gaps = 12/261 (4%)
Frame = +2
Query: 419 YLLNLLMKINAKLGGLNSLLQVEASSSIPHVSQVPTIILGMDVSHGHPGQDR-PSVAAVV 595
YL NL +K+N K+GG N++L+ IP ++ PTI+ G DV+H PG+D PS+AAVV
Sbjct: 742 YLENLALKMNVKVGGRNTVLEDALHKKIPILTDRPTIVFGADVTHPSPGEDASPSIAAVV 801
Query: 596 SSRQWPLISRYRASVHTQSARLEMMSSLFKP-----RGTDDDGLIRESLIDFYTSSGKRK 760
+S WP +++Y+ V TQS R E++S+L+ +G G+IRE L FY +G+ K
Sbjct: 802 ASMDWPEVTKYKCLVSTQSHREEIISNLYTEVKDPLKGIIRGGMIRELLRSFYQETGQ-K 860
Query: 761 PEHIIIFRDGVSESQFTQVINIELDQIIEACKFLDEKWSPKFTVIVAQKNHHTKFFQTAS 940
P II +RDG+SE QF+QV+ E+D I +AC L E + P T +V QK HHT+ F
Sbjct: 861 PSRIIFYRDGISEGQFSQVLLYEMDAIRKACASLQEGYLPPVTFVVVQKRHHTRLFPENR 920
Query: 941 PD------NVLPGTVVDSKVCHPKNFDFYMCAHAGMIGTTRPTHYHVLHDEIGFSADEMQ 1102
D N+LPGTVVD+ +CHP FDFY+C+H+G+ GT+RPTHYHVL DE GF AD +Q
Sbjct: 921 RDMMDRSGNILPGTVVDTMICHPSEFDFYLCSHSGIKGTSRPTHYHVLLDENGFKADTLQ 980
Query: 1103 EFVHSLSYVYTIADRWIEIAP 1165
++LSY Y R + I P
Sbjct: 981 TLTYNLSYTYARCTRAVSIVP 1001
>sptrnew|AAN41341|AAN41341 Putative leaf development protein
Argonaute.
Length = 1048
Score = 238 bits (608), Expect = 1e-61
Identities = 127/263 (48%), Positives = 165/263 (62%), Gaps = 14/263 (5%)
Frame = +2
Query: 419 YLLNLLMKINAKLGGLNSLLQVEASSSIPHVSQVPTIILGMDVSHGHPGQDR-PSVAAVV 595
Y+ N+ +KIN K+GG N++L S IP VS PTII G DV+H HPG+D PS+AAVV
Sbjct: 719 YMANVALKINVKVGGRNTVLVDALSRRIPLVSDRPTIIFGADVTHPHPGEDSSPSIAAVV 778
Query: 596 SSRQWPLISRYRASVHTQSARLEMMSSLFKP-----RGTDDDGLIRESLIDFYTSSGKRK 760
+S+ WP I++Y V Q+ R E++ LFK +G G+I+E LI F S+G K
Sbjct: 779 ASQDWPEITKYAGLVCAQAHRQELIQDLFKEWKDPQKGVVTGGMIKELLIAFRRSTG-HK 837
Query: 761 PEHIIIFRDGVSESQFTQVINIELDQIIEACKFLDEKWSPKFTVIVAQKNHHTKFFQTAS 940
P II +RDGVSE QF QV+ ELD I +AC L+ + P T +V QK HHT+ F
Sbjct: 838 PLRIIFYRDGVSEGQFYQVLLYELDAIRKACASLEAGYQPPVTFVVVQKRHHTRLFAQNH 897
Query: 941 PD--------NVLPGTVVDSKVCHPKNFDFYMCAHAGMIGTTRPTHYHVLHDEIGFSADE 1096
D N+LPGTVVDSK+CHP FDFY+C+HAG+ GT+RP HYHVL DE F+AD
Sbjct: 898 NDRHSVDRSGNILPGTVVDSKICHPTEFDFYLCSHAGIQGTSRPAHYHVLWDENNFTADG 957
Query: 1097 MQEFVHSLSYVYTIADRWIEIAP 1165
+Q ++L Y Y R + I P
Sbjct: 958 LQSLTNNLCYTYARCTRSVSIVP 980
>sw|O04379|AGO1_ARATH Argonaute protein.
Length = 1048
Score = 238 bits (608), Expect = 1e-61
Identities = 127/263 (48%), Positives = 165/263 (62%), Gaps = 14/263 (5%)
Frame = +2
Query: 419 YLLNLLMKINAKLGGLNSLLQVEASSSIPHVSQVPTIILGMDVSHGHPGQDR-PSVAAVV 595
Y+ N+ +KIN K+GG N++L S IP VS PTII G DV+H HPG+D PS+AAVV
Sbjct: 719 YMANVALKINVKVGGRNTVLVDALSRRIPLVSDRPTIIFGADVTHPHPGEDSSPSIAAVV 778
Query: 596 SSRQWPLISRYRASVHTQSARLEMMSSLFKP-----RGTDDDGLIRESLIDFYTSSGKRK 760
+S+ WP I++Y V Q+ R E++ LFK +G G+I+E LI F S+G K
Sbjct: 779 ASQDWPEITKYAGLVCAQAHRQELIQDLFKEWKDPQKGVVTGGMIKELLIAFRRSTG-HK 837
Query: 761 PEHIIIFRDGVSESQFTQVINIELDQIIEACKFLDEKWSPKFTVIVAQKNHHTKFFQTAS 940
P II +RDGVSE QF QV+ ELD I +AC L+ + P T +V QK HHT+ F
Sbjct: 838 PLRIIFYRDGVSEGQFYQVLLYELDAIRKACASLEAGYQPPVTFVVVQKRHHTRLFAQNH 897
Query: 941 PD--------NVLPGTVVDSKVCHPKNFDFYMCAHAGMIGTTRPTHYHVLHDEIGFSADE 1096
D N+LPGTVVDSK+CHP FDFY+C+HAG+ GT+RP HYHVL DE F+AD
Sbjct: 898 NDRHSVDRSGNILPGTVVDSKICHPTEFDFYLCSHAGIQGTSRPAHYHVLWDENNFTADG 957
Query: 1097 MQEFVHSLSYVYTIADRWIEIAP 1165
+Q ++L Y Y R + I P
Sbjct: 958 LQSLTNNLCYTYARCTRSVSIVP 980
>sw|Q9XGW1|PINH_ARATH PINHEAD protein (ZWILLE protein).
Length = 988
Score = 235 bits (600), Expect = 9e-61
Identities = 124/263 (47%), Positives = 166/263 (63%), Gaps = 14/263 (5%)
Frame = +2
Query: 419 YLLNLLMKINAKLGGLNSLLQVEASSSIPHVSQVPTIILGMDVSHGHPGQDR-PSVAAVV 595
YL N+ +KIN K+GG N++L S IP VS +PTII G DV+H G++ PS+AAVV
Sbjct: 668 YLANVSLKINVKMGGRNTVLVDAISCRIPLVSDIPTIIFGADVTHPENGEESSPSIAAVV 727
Query: 596 SSRQWPLISRYRASVHTQSARLEMMSSLFKP-----RGTDDDGLIRESLIDFYTSSGKRK 760
+S+ WP +++Y V Q+ R E++ L+K RGT G+IR+ LI F ++G+ K
Sbjct: 728 ASQDWPEVTKYAGLVCAQAHRQELIQDLYKTWQDPVRGTVSGGMIRDLLISFRKATGQ-K 786
Query: 761 PEHIIIFRDGVSESQFTQVINIELDQIIEACKFLDEKWSPKFTVIVAQKNHHTKFFQTAS 940
P II +RDGVSE QF QV+ ELD I +AC L+ + P T IV QK HHT+ F
Sbjct: 787 PLRIIFYRDGVSEGQFYQVLLYELDAIRKACASLEPNYQPPVTFIVVQKRHHTRLFANNH 846
Query: 941 PD--------NVLPGTVVDSKVCHPKNFDFYMCAHAGMIGTTRPTHYHVLHDEIGFSADE 1096
D N+LPGTVVD+K+CHP FDFY+C+HAG+ GT+RP HYHVL DE F+AD
Sbjct: 847 RDKNSTDRSGNILPGTVVDTKICHPTEFDFYLCSHAGIQGTSRPAHYHVLWDENNFTADG 906
Query: 1097 MQEFVHSLSYVYTIADRWIEIAP 1165
+Q ++L Y Y R + I P
Sbjct: 907 IQSLTNNLCYTYARCTRSVSIVP 929
>sptr|Q851R2|Q851R2 Putative argonaute protein.
Length = 1058
Score = 233 bits (595), Expect = 4e-60
Identities = 124/263 (47%), Positives = 167/263 (63%), Gaps = 14/263 (5%)
Frame = +2
Query: 419 YLLNLLMKINAKLGGLNSLLQ-VEASSSIPHVSQVPTIILGMDVSHGHPGQDRPS-VAAV 592
YL N+ +KIN K+GG N++L+ + IP VS+VPTII G DV+H PG+D S +AAV
Sbjct: 738 YLENVALKINVKVGGRNTVLERAFIRNGIPFVSEVPTIIFGADVTHPPPGEDSASSIAAV 797
Query: 593 VSSRQWPLISRYRASVHTQSARLEMMSSLFK----PRGTDDDGLIRESLIDFYTSSGKRK 760
V+S WP I++YR V Q R E++ LF P + G+IRE LI F +G+R
Sbjct: 798 VASMDWPEITKYRGLVSAQPHRQEIIEDLFSVGKDPVKVVNGGMIRELLIAFRKKTGRR- 856
Query: 761 PEHIIIFRDGVSESQFTQVINIELDQIIEACKFLDEKWSPKFTVIVAQKNHHTKFF---- 928
PE II +RDGVSE QF+ V+ E+D I +AC L+E + P T +V QK HHT+ F
Sbjct: 857 PERIIFYRDGVSEGQFSHVLLHEMDAIRKACASLEEGYLPPVTFVVVQKRHHTRLFPEVH 916
Query: 929 ----QTASPDNVLPGTVVDSKVCHPKNFDFYMCAHAGMIGTTRPTHYHVLHDEIGFSADE 1096
T N+LPGTVVD ++CHP FDFY+C+HAG+ GT+RPTHYHVL+DE F+AD
Sbjct: 917 GRRDMTDKSGNILPGTVVDRQICHPTEFDFYLCSHAGIQGTSRPTHYHVLYDENHFTADA 976
Query: 1097 MQEFVHSLSYVYTIADRWIEIAP 1165
+Q ++L Y Y R + + P
Sbjct: 977 LQSLTNNLCYTYARCTRAVSVVP 999
>sw|Q9SJK3|AGOL_ARATH Argonaute-like protein At2g27880.
Length = 997
Score = 227 bits (579), Expect = 3e-58
Identities = 118/263 (44%), Positives = 165/263 (62%), Gaps = 14/263 (5%)
Frame = +2
Query: 419 YLLNLLMKINAKLGGLNSLLQVEASSSIPHVSQVPTIILGMDVSHGHPGQDR-PSVAAVV 595
Y+ N+ +KIN K GG N++L +IP ++ PTII+G DV+H PG+D PS+AAVV
Sbjct: 680 YMENVALKINVKTGGRNTVLNDAIRRNIPLITDRPTIIMGADVTHPQPGEDSSPSIAAVV 739
Query: 596 SSRQWPLISRYRASVHTQSARLEMMSSLFK-----PRGTDDDGLIRESLIDFYTSSGKRK 760
+S WP I++YR V Q+ R E++ L+K RG GLIRE I F ++G+
Sbjct: 740 ASMDWPEINKYRGLVSAQAHREEIIQDLYKLVQDPQRGLVHSGLIREHFIAFRRATGQI- 798
Query: 761 PEHIIIFRDGVSESQFTQVINIELDQIIEACKFLDEKWSPKFTVIVAQKNHHTKFF---- 928
P+ II +RDGVSE QF+QV+ E+ I +AC L E + P+ T ++ QK HHT+ F
Sbjct: 799 PQRIIFYRDGVSEGQFSQVLLHEMTAIRKACNSLQENYVPRVTFVIVQKRHHTRLFPEQH 858
Query: 929 ----QTASPDNVLPGTVVDSKVCHPKNFDFYMCAHAGMIGTTRPTHYHVLHDEIGFSADE 1096
T N+ PGTVVD+K+CHP FDFY+ +HAG+ GT+RP HYHVL DE GF+AD+
Sbjct: 859 GNRDMTDKSGNIQPGTVVDTKICHPNEFDFYLNSHAGIQGTSRPAHYHVLLDENGFTADQ 918
Query: 1097 MQEFVHSLSYVYTIADRWIEIAP 1165
+Q ++L Y Y + + I P
Sbjct: 919 LQMLTNNLCYTYARCTKSVSIVP 941
>sptr|Q9LP83|Q9LP83 T1N15.2.
Length = 1123
Score = 224 bits (571), Expect = 2e-57
Identities = 127/278 (45%), Positives = 165/278 (59%), Gaps = 29/278 (10%)
Frame = +2
Query: 419 YLLNLLMKINAKLGGLNSLLQVEASSSIPHVSQVPTIILGMDVSHGHPGQDR-PSVAAVV 595
Y+ N+ +KIN K+GG N++L S IP VS PTII G DV+H HPG+D PS+AAVV
Sbjct: 779 YMANVALKINVKVGGRNTVLVDALSRRIPLVSDRPTIIFGADVTHPHPGEDSSPSIAAVV 838
Query: 596 SSRQWPLISRYRASVHTQSARLEMMSSLFKP-----RGTDDDGLIR------------ES 724
+S+ WP I++Y V Q+ R E++ LFK +G G+I+ E
Sbjct: 839 ASQDWPEITKYAGLVCAQAHRQELIQDLFKEWKDPQKGVVTGGMIKYVWMLFNIFVIGEL 898
Query: 725 LIDFYTSSGKRKPEHIIIFRDGVSESQFTQVINIELDQIIEACKFLDEKWSPKFTVIVAQ 904
LI F S+G KP II +RDGVSE QF QV+ ELD I +AC L+ + P T +V Q
Sbjct: 899 LIAFRRSTG-HKPLRIIFYRDGVSEGQFYQVLLYELDAIRKACASLEAGYQPPVTFVVVQ 957
Query: 905 KNHHTKFFQTASPD--------NVLPG---TVVDSKVCHPKNFDFYMCAHAGMIGTTRPT 1051
K HHT+ F D N+LPG TVVDSK+CHP FDFY+C+HAG+ GT+RP
Sbjct: 958 KRHHTRLFAQNHNDRHSVDRSGNILPGETSTVVDSKICHPTEFDFYLCSHAGIQGTSRPA 1017
Query: 1052 HYHVLHDEIGFSADEMQEFVHSLSYVYTIADRWIEIAP 1165
HYHVL DE F+AD +Q ++L Y Y R + I P
Sbjct: 1018 HYHVLWDENNFTADGLQSLTNNLCYTYARCTRSVSIVP 1055
>sptrnew|BAC83909|BAC83909 Putative leaf development protein
Argonaute.
Length = 1052
Score = 216 bits (551), Expect = 5e-55
Identities = 119/276 (43%), Positives = 161/276 (58%), Gaps = 27/276 (9%)
Frame = +2
Query: 419 YLLNLLMKINAKLGGLNSLLQVEASSSIPHVSQVPTIILGMDVSHGHPGQDRP-SVAAVV 595
+L N+ +KIN K GG NS+LQ +P + TII G DV+H G+D S+AAVV
Sbjct: 720 FLENVSLKINVKAGGRNSVLQ---RPLVPGGLENTTIIFGADVTHPASGEDSSASIAAVV 776
Query: 596 SSRQWPLISRYRASVHTQSARLEMMSSLF--------------KPRGTDDD----GLIRE 721
+S WP I++Y+A V Q R E++ LF K G+ + G+ RE
Sbjct: 777 ASMDWPEITKYKALVSAQPPRQEIIQDLFTMTEVAQNADAPAQKAEGSKKNFICGGMFRE 836
Query: 722 SLIDFYTSSGKRKPEHIIIFRDGVSESQFTQVINIELDQIIEACKFLDEKWSPKFTVIVA 901
L+ FY+ + KRKP+ II +RDGVS+ QF V+ E+D I +A LD + P T +V
Sbjct: 837 LLMSFYSKNAKRKPQRIIFYRDGVSDGQFLHVLLYEMDAIKKAIASLDPAYRPLVTFVVV 896
Query: 902 QKNHHTKFFQ--------TASPDNVLPGTVVDSKVCHPKNFDFYMCAHAGMIGTTRPTHY 1057
QK HHT+ F T NV PGTVVD+ +CHP FDFY+C+HAG+ GT+RPTHY
Sbjct: 897 QKRHHTRLFPEVHGRQDLTDRSGNVRPGTVVDTNICHPSEFDFYLCSHAGIQGTSRPTHY 956
Query: 1058 HVLHDEIGFSADEMQEFVHSLSYVYTIADRWIEIAP 1165
HVLHDE FSAD++Q ++L Y Y R + + P
Sbjct: 957 HVLHDENRFSADQLQMLTYNLCYTYARCTRSVSVVP 992
>sptr|Q7XZZ9|Q7XZZ9 Putative leaf development and shoot apical
meristem regulating protein.
Length = 895
Score = 207 bits (528), Expect = 2e-52
Identities = 106/236 (44%), Positives = 148/236 (62%), Gaps = 14/236 (5%)
Frame = +2
Query: 500 IPHVSQVPTIILGMDVSHGHPGQDR-PSVAAVVSSRQWPLISRYRASVHTQSARLEMMSS 676
IP ++ +PT+I G DV+H G+D PS+AAVV+S WP +S+Y+ SV +QS R E+++
Sbjct: 599 IPLLTDMPTMIFGADVTHPPAGEDSSPSIAAVVASMDWPEVSKYKCSVSSQSHREEIIAD 658
Query: 677 LFKPRGTDDD-----GLIRESLIDFYTSSGKRKPEHIIIFRDGVSESQFTQVINIELDQI 841
LF + G+IRE + F ++G KP II +RDGVSE QF+QV+ E+D I
Sbjct: 659 LFTEVKDSQNRLVYGGMIRELIESFRKANGSYKPGRIIFYRDGVSEGQFSQVLLSEMDAI 718
Query: 842 IEACKFLDEKWSPKFTVIVAQKNHHTKFF--------QTASPDNVLPGTVVDSKVCHPKN 997
+AC ++E + P T +V QK HHT+ F Q N+LPGTVVD+K+CHP
Sbjct: 719 RKACASIEEGYLPPVTFVVVQKRHHTRLFPEDHHARDQMDRSRNILPGTVVDTKICHPSE 778
Query: 998 FDFYMCAHAGMIGTTRPTHYHVLHDEIGFSADEMQEFVHSLSYVYTIADRWIEIAP 1165
FDFY+C+H+G+ GT+ PTHY+VL DE FSAD +Q + L Y Y R + I P
Sbjct: 779 FDFYLCSHSGIQGTSHPTHYYVLFDENNFSADALQTLTYHLCYTYARCTRSVSIVP 834
>sptrnew|AAQ92355|AAQ92355 ZIPPY.
Length = 990
Score = 201 bits (510), Expect = 3e-50
Identities = 115/255 (45%), Positives = 158/255 (61%), Gaps = 6/255 (2%)
Frame = +2
Query: 419 YLLNLLMKINAKLGGLNSLLQVEASSSIPHVSQV--PTIILGMDVSHGHPGQD-RPSVAA 589
++ NL +KINAK+GG + L S IP + + P I +G DV+H HP D PSVAA
Sbjct: 691 FVSNLALKINAKIGGSMTELYNSIPSHIPRLLRPDEPVIFMGADVTHPHPFDDCSPSVAA 750
Query: 590 VVSSRQWPLISRYRASVHTQSARLEMMSSLFKPRGTDDDGLIRESLIDFYTSSGKRKPEH 769
VV S WP +RY + + +Q+ R E++ L D +++E L DFY + K+ P
Sbjct: 751 VVGSINWPEANRYVSRMRSQTHRQEIIQDL--------DLMVKELLDDFYKAV-KKLPNR 801
Query: 770 IIIFRDGVSESQFTQVINIELDQIIEAC-KFLDEKWSPKFTVIVAQKNHHTKFFQTASPD 946
II FRDGVSE+QF +V+ EL I AC KF D ++P T V QK HHT+ F+ PD
Sbjct: 802 IIFFRDGVSETQFKKVLQEELQSIKTACSKFQD--YNPSITFAVVQKRHHTRLFR-CDPD 858
Query: 947 --NVLPGTVVDSKVCHPKNFDFYMCAHAGMIGTTRPTHYHVLHDEIGFSADEMQEFVHSL 1120
N+ PGTVVD+ + HPK FDFY+C+H G+ GT+RPTHYH+L DE F++DE+Q V++L
Sbjct: 859 HENIPPGTVVDTVITHPKEFDFYLCSHLGVKGTSRPTHYHILWDENEFTSDELQRLVYNL 918
Query: 1121 SYVYTIADRWIEIAP 1165
Y + + I I P
Sbjct: 919 CYTFVRCTKPISIVP 933
>sptr|Q9C793|Q9C793 Pinhead-like protein.
Length = 990
Score = 201 bits (510), Expect = 3e-50
Identities = 115/255 (45%), Positives = 158/255 (61%), Gaps = 6/255 (2%)
Frame = +2
Query: 419 YLLNLLMKINAKLGGLNSLLQVEASSSIPHVSQV--PTIILGMDVSHGHPGQD-RPSVAA 589
++ NL +KINAK+GG + L S IP + + P I +G DV+H HP D PSVAA
Sbjct: 691 FVSNLALKINAKIGGSMTELYNSIPSHIPRLLRPDEPVIFMGADVTHPHPFDDCSPSVAA 750
Query: 590 VVSSRQWPLISRYRASVHTQSARLEMMSSLFKPRGTDDDGLIRESLIDFYTSSGKRKPEH 769
VV S WP +RY + + +Q+ R E++ L D +++E L DFY + K+ P
Sbjct: 751 VVGSINWPEANRYVSRMRSQTHRQEIIQDL--------DLMVKELLDDFYKAV-KKLPNR 801
Query: 770 IIIFRDGVSESQFTQVINIELDQIIEAC-KFLDEKWSPKFTVIVAQKNHHTKFFQTASPD 946
II FRDGVSE+QF +V+ EL I AC KF D ++P T V QK HHT+ F+ PD
Sbjct: 802 IIFFRDGVSETQFKKVLQEELQSIKTACSKFQD--YNPSITFAVVQKRHHTRLFR-CDPD 858
Query: 947 --NVLPGTVVDSKVCHPKNFDFYMCAHAGMIGTTRPTHYHVLHDEIGFSADEMQEFVHSL 1120
N+ PGTVVD+ + HPK FDFY+C+H G+ GT+RPTHYH+L DE F++DE+Q V++L
Sbjct: 859 HENIPPGTVVDTVITHPKEFDFYLCSHLGVKGTSRPTHYHILWDENEFTSDELQRLVYNL 918
Query: 1121 SYVYTIADRWIEIAP 1165
Y + + I I P
Sbjct: 919 CYTFVRCTKPISIVP 933
>sptr|Q852N2|Q852N2 Putative argonaute protein.
Length = 1192
Score = 195 bits (496), Expect = 1e-48
Identities = 111/262 (42%), Positives = 153/262 (58%), Gaps = 13/262 (4%)
Frame = +2
Query: 419 YLLNLLMKINAKLGGLNSLLQVEASSSIPHVSQVPTIILGMDVSHGHPGQDRPS-VAAVV 595
YL N+ +KIN K SQ +++L +SH PG+D S +AAVV
Sbjct: 514 YLENVALKINVKK------------------SQQSSLVL---MSHTPPGEDSASSIAAVV 552
Query: 596 SSRQWPLISRYRASVHTQSARLEMMSSLFK----PRGTDDDGLIRESLIDFYTSSGKRKP 763
+S WP I++YR V QS R E++ LF P + G+IRE LI F +G+R P
Sbjct: 553 ASMDWPEITKYRGLVSAQSHRQEIIEDLFSVGKDPVKVVNGGMIREFLIAFRKKTGRR-P 611
Query: 764 EHIIIFRDGVSESQFTQVINIELDQIIEACKFLDEKWSPKFTVIVAQKNHHTKFF----- 928
E II +RDGVSE QF++V+ E+D I +AC L+E + P T +V QK HHT+ F
Sbjct: 612 ERIIFYRDGVSEGQFSRVLLHEMDAIRKACASLEEGYLPPVTFVVVQKRHHTRLFPEVHG 671
Query: 929 ---QTASPDNVLPGTVVDSKVCHPKNFDFYMCAHAGMIGTTRPTHYHVLHDEIGFSADEM 1099
T N+LPGTV D ++CHP F FY+C+HAG+ GT+RPTHYHVL+DE F+ADE+
Sbjct: 672 RRDMTDKSGNILPGTVKDRQICHPTEFYFYLCSHAGIQGTSRPTHYHVLYDENHFTADEL 731
Query: 1100 QEFVHSLSYVYTIADRWIEIAP 1165
Q ++L Y+Y + + P
Sbjct: 732 QTLTNNLCYIYARCTHAVSVVP 753
>sptr|Q8T059|Q8T059 LD26301p.
Length = 424
Score = 191 bits (485), Expect = 2e-47
Identities = 111/253 (43%), Positives = 146/253 (57%), Gaps = 7/253 (2%)
Frame = +2
Query: 422 LLNLLMKINAKLGGLNSLLQVEASSSIPHVSQVPTIILGMDVSHGHPGQDR-PSVAAVVS 598
L NL +KIN KLGG+NS+L S+ P V P I LG DV+H G ++ PS+AAVV
Sbjct: 124 LSNLCLKINVKLGGINSIL---VPSTRPKVFNEPVIFLGADVTHPPAGDNKKPSIAAVVG 180
Query: 599 SRQWPLISRYRASVHTQSARLEMMSSLFKPRGTDDDGLIRESLIDFYTSSGKRKPEHIII 778
S SRY A+V Q R E++ L ++RE LI FY S+G KP II+
Sbjct: 181 SMD-AHPSRYAATVRVQQHRQEIIQEL--------SSMVRELLIMFYKSTGGYKPHRIIL 231
Query: 779 FRDGVSESQFTQVINIELDQIIEACKFLDEKWSPKFTVIVAQKNHHTKFF------QTAS 940
+RDGVSE QF V+ EL I EAC L+ ++ P T IV QK HHT+ F Q+
Sbjct: 232 YRDGVSEGQFPHVLQHELTAIREACIKLEPEYRPGITFIVVQKRHHTRLFCAEKKEQSGK 291
Query: 941 PDNVLPGTVVDSKVCHPKNFDFYMCAHAGMIGTTRPTHYHVLHDEIGFSADEMQEFVHSL 1120
N+ GT VD + HP FDFY+C+H G+ GT+RP+HYHVL D+ F +DE+Q + L
Sbjct: 292 SGNIPAGTTVDVGITHPTEFDFYLCSHQGIQGTSRPSHYHVLWDDNHFDSDELQCLTYQL 351
Query: 1121 SYVYTIADRWIEI 1159
+ Y R + I
Sbjct: 352 CHTYVRCTRSVSI 364
>sptr|Q9V3L2|Q9V3L2 AGO1 protein.
Length = 950
Score = 190 bits (482), Expect = 5e-47
Identities = 111/253 (43%), Positives = 145/253 (57%), Gaps = 7/253 (2%)
Frame = +2
Query: 422 LLNLLMKINAKLGGLNSLLQVEASSSIPHVSQVPTIILGMDVSHGHPGQDR-PSVAAVVS 598
L NL +KIN KLGG+NS+L S P V P I LG DV+H G ++ PS+AAVV
Sbjct: 650 LSNLCLKINVKLGGINSIL---VPSIRPKVFNEPVIFLGADVTHPPAGDNKKPSIAAVVG 706
Query: 599 SRQWPLISRYRASVHTQSARLEMMSSLFKPRGTDDDGLIRESLIDFYTSSGKRKPEHIII 778
S SRY A+V Q R E++ L ++RE LI FY S+G KP II+
Sbjct: 707 SMD-AHPSRYAATVRVQQHRQEIIQEL--------SSMVRELLIMFYKSTGGYKPHRIIL 757
Query: 779 FRDGVSESQFTQVINIELDQIIEACKFLDEKWSPKFTVIVAQKNHHTKFF------QTAS 940
+RDGVSE QF V+ EL I EAC L+ ++ P T IV QK HHT+ F Q+
Sbjct: 758 YRDGVSEGQFPHVLQHELTAIREACIKLEPEYRPGITFIVVQKRHHTRLFCAEKKEQSGK 817
Query: 941 PDNVLPGTVVDSKVCHPKNFDFYMCAHAGMIGTTRPTHYHVLHDEIGFSADEMQEFVHSL 1120
N+ GT VD + HP FDFY+C+H G+ GT+RP+HYHVL D+ F +DE+Q + L
Sbjct: 818 SGNIPAGTTVDVGITHPTEFDFYLCSHQGIQGTSRPSHYHVLWDDNHFDSDELQCLTYQL 877
Query: 1121 SYVYTIADRWIEI 1159
+ Y R + I
Sbjct: 878 CHTYVRCTRSVSI 890
>sptr|Q960T1|Q960T1 LD36719p.
Length = 601
Score = 190 bits (482), Expect = 5e-47
Identities = 111/253 (43%), Positives = 145/253 (57%), Gaps = 7/253 (2%)
Frame = +2
Query: 422 LLNLLMKINAKLGGLNSLLQVEASSSIPHVSQVPTIILGMDVSHGHPGQDR-PSVAAVVS 598
L NL +KIN KLGG+NS+L S P V P I LG DV+H G ++ PS+AAVV
Sbjct: 301 LSNLCLKINVKLGGINSIL---VPSIRPKVFNEPVIFLGADVTHPPAGDNKKPSIAAVVG 357
Query: 599 SRQWPLISRYRASVHTQSARLEMMSSLFKPRGTDDDGLIRESLIDFYTSSGKRKPEHIII 778
S SRY A+V Q R E++ L ++RE LI FY S+G KP II+
Sbjct: 358 SMD-AHPSRYAATVRVQQHRQEIIQEL--------SSMVRELLIMFYKSTGGYKPHRIIL 408
Query: 779 FRDGVSESQFTQVINIELDQIIEACKFLDEKWSPKFTVIVAQKNHHTKFF------QTAS 940
+RDGVSE QF V+ EL I EAC L+ ++ P T IV QK HHT+ F Q+
Sbjct: 409 YRDGVSEGQFPHVLQHELTAIREACIKLEPEYRPGITFIVVQKRHHTRLFCAEKKEQSGK 468
Query: 941 PDNVLPGTVVDSKVCHPKNFDFYMCAHAGMIGTTRPTHYHVLHDEIGFSADEMQEFVHSL 1120
N+ GT VD + HP FDFY+C+H G+ GT+RP+HYHVL D+ F +DE+Q + L
Sbjct: 469 SGNIPAGTTVDVGITHPTEFDFYLCSHQGIQGTSRPSHYHVLWDDNHFDSDELQCLTYQL 528
Query: 1121 SYVYTIADRWIEI 1159
+ Y R + I
Sbjct: 529 CHTYVRCTRSVSI 541
>sptr|Q9V6V6|Q9V6V6 AGO1 protein.
Length = 984
Score = 190 bits (482), Expect = 5e-47
Identities = 111/253 (43%), Positives = 145/253 (57%), Gaps = 7/253 (2%)
Frame = +2
Query: 422 LLNLLMKINAKLGGLNSLLQVEASSSIPHVSQVPTIILGMDVSHGHPGQDR-PSVAAVVS 598
L NL +KIN KLGG+NS+L S P V P I LG DV+H G ++ PS+AAVV
Sbjct: 684 LSNLCLKINVKLGGINSIL---VPSIRPKVFNEPVIFLGADVTHPPAGDNKKPSIAAVVG 740
Query: 599 SRQWPLISRYRASVHTQSARLEMMSSLFKPRGTDDDGLIRESLIDFYTSSGKRKPEHIII 778
S SRY A+V Q R E++ L ++RE LI FY S+G KP II+
Sbjct: 741 SMD-AHPSRYAATVRVQQHRQEIIQEL--------SSMVRELLIMFYKSTGGYKPHRIIL 791
Query: 779 FRDGVSESQFTQVINIELDQIIEACKFLDEKWSPKFTVIVAQKNHHTKFF------QTAS 940
+RDGVSE QF V+ EL I EAC L+ ++ P T IV QK HHT+ F Q+
Sbjct: 792 YRDGVSEGQFPHVLQHELTAIREACIKLEPEYRPGITFIVVQKRHHTRLFCAEKKEQSGK 851
Query: 941 PDNVLPGTVVDSKVCHPKNFDFYMCAHAGMIGTTRPTHYHVLHDEIGFSADEMQEFVHSL 1120
N+ GT VD + HP FDFY+C+H G+ GT+RP+HYHVL D+ F +DE+Q + L
Sbjct: 852 SGNIPAGTTVDVGITHPTEFDFYLCSHQGIQGTSRPSHYHVLWDDNHFDSDELQCLTYQL 911
Query: 1121 SYVYTIADRWIEI 1159
+ Y R + I
Sbjct: 912 CHTYVRCTRSVSI 924
>sptrnew|EAA00062|EAA00062 EbiP8896 (Fragment).
Length = 922
Score = 189 bits (480), Expect = 8e-47
Identities = 111/253 (43%), Positives = 144/253 (56%), Gaps = 7/253 (2%)
Frame = +2
Query: 422 LLNLLMKINAKLGGLNSLLQVEASSSIPHVSQVPTIILGMDVSHGHPGQDR-PSVAAVVS 598
L NL +KIN KLGG+NS+L S P V P I LG DV+H G ++ PS+AAVV
Sbjct: 622 LSNLCLKINVKLGGINSIL---VPSIRPKVFDEPVIFLGADVTHPPAGDNKKPSIAAVVG 678
Query: 599 SRQWPLISRYRASVHTQSARLEMMSSLFKPRGTDDDGLIRESLIDFYTSSGKRKPEHIII 778
S SRY A+V Q R E++ L ++RE LI FY S+G KP II+
Sbjct: 679 SMD-AHPSRYAATVRVQQHRQEIIQEL--------SSMVRELLIMFYKSTGGFKPHRIIL 729
Query: 779 FRDGVSESQFTQVINIELDQIIEACKFLDEKWSPKFTVIVAQKNHHTKFF------QTAS 940
+RDGVSE QF V+ EL I EAC L+ + P T IV QK HHT+ F Q+
Sbjct: 730 YRDGVSEGQFPHVLQHELTAIREACIKLEADYKPGITFIVVQKRHHTRLFCADKKEQSGK 789
Query: 941 PDNVLPGTVVDSKVCHPKNFDFYMCAHAGMIGTTRPTHYHVLHDEIGFSADEMQEFVHSL 1120
N+ GT VD + HP FDFY+C+H G+ GT+RP+HYHVL D+ F +DE+Q + L
Sbjct: 790 SGNIPAGTTVDVGITHPTEFDFYLCSHQGIQGTSRPSHYHVLWDDNHFESDELQCLTYQL 849
Query: 1121 SYVYTIADRWIEI 1159
+ Y R + I
Sbjct: 850 CHTYVRCTRSVSI 862
>sw|Q9QZ81|I2C2_RAT Eukaryotic translation initiation factor 2C 2
(eIF2C 2) (eIF-2C 2) (Golgi ER protein 95 kDa) (GERp95).
Length = 863
Score = 189 bits (479), Expect = 1e-46
Identities = 112/253 (44%), Positives = 151/253 (59%), Gaps = 7/253 (2%)
Frame = +2
Query: 422 LLNLLMKINAKLGGLNSLLQVEASSSIPHVSQVPTIILGMDVSHGHPGQDR-PSVAAVVS 598
L NL +KIN KLGG+N++L + P V Q P I LG DV+H G + PS+AAVV
Sbjct: 564 LSNLCLKINVKLGGVNNILLPQGR---PPVFQQPVIFLGADVTHPPAGDGKKPSIAAVVG 620
Query: 599 SRQWPLISRYRASVHTQSARLEMMSSLFKPRGTDDDGLIRESLIDFYTSSGKRKPEHIII 778
S +RY A+V Q R E++ L ++RE LI FY S+ + KP II
Sbjct: 621 SMD-AHPNRYCATVRVQQHRQEIIQDLA--------AMVRELLIQFYKST-RFKPTRIIF 670
Query: 779 FRDGVSESQFTQVINIELDQIIEACKFLDEKWSPKFTVIVAQKNHHTKFFQTASPD---- 946
+RDGVSE QF QV++ EL I EAC L++++ P T IV QK HHT+ F T +
Sbjct: 671 YRDGVSEGQFQQVLHHELLAIREACIKLEKEYQPGITFIVVQKRHHTRLFCTDKNERVGK 730
Query: 947 --NVLPGTVVDSKVCHPKNFDFYMCAHAGMIGTTRPTHYHVLHDEIGFSADEMQEFVHSL 1120
N+ GT VD+K+ HP FDFY+C+HAG+ GT+RP+HYHVL D+ FS+DE+Q + L
Sbjct: 731 SGNIPAGTTVDTKITHPTEFDFYLCSHAGIQGTSRPSHYHVLWDDNRFSSDELQILTYQL 790
Query: 1121 SYVYTIADRWIEI 1159
+ Y R + I
Sbjct: 791 CHTYVRCTRSVSI 803
>sptr|Q8R3Q7|Q8R3Q7 Similar to eukaryotic translation initiation
factor 2C, 2 (Fragment).
Length = 530
Score = 188 bits (477), Expect = 2e-46
Identities = 112/253 (44%), Positives = 150/253 (59%), Gaps = 7/253 (2%)
Frame = +2
Query: 422 LLNLLMKINAKLGGLNSLLQVEASSSIPHVSQVPTIILGMDVSHGHPGQDR-PSVAAVVS 598
L NL +KIN KLGG+N++L + P V Q P I LG DV+H G + PS+AAVV
Sbjct: 231 LSNLCLKINVKLGGVNNILLPQGR---PPVFQQPVIFLGADVTHPPAGDGKKPSIAAVVG 287
Query: 599 SRQWPLISRYRASVHTQSARLEMMSSLFKPRGTDDDGLIRESLIDFYTSSGKRKPEHIII 778
S +RY A+V Q R E++ L ++RE LI FY S+ + KP II
Sbjct: 288 SMD-AHPNRYCATVRVQQHRQEIIQDLA--------AMVRELLIQFYKST-RFKPTRIIF 337
Query: 779 FRDGVSESQFTQVINIELDQIIEACKFLDEKWSPKFTVIVAQKNHHTKFFQTASPD---- 946
+RDGVSE QF QV++ EL I EAC L++ + P T IV QK HHT+ F T +
Sbjct: 338 YRDGVSEGQFQQVLHHELLAIREACIKLEKDYQPGITFIVVQKRHHTRLFCTDKNERVGK 397
Query: 947 --NVLPGTVVDSKVCHPKNFDFYMCAHAGMIGTTRPTHYHVLHDEIGFSADEMQEFVHSL 1120
N+ GT VD+K+ HP FDFY+C+HAG+ GT+RP+HYHVL D+ FS+DE+Q + L
Sbjct: 398 SGNIPAGTTVDTKITHPTEFDFYLCSHAGIQGTSRPSHYHVLWDDNRFSSDELQILTYQL 457
Query: 1121 SYVYTIADRWIEI 1159
+ Y R + I
Sbjct: 458 CHTYVRCTRSVSI 470
>sptrnew|AAH56639|AAH56639 Hypothetical protein (Fragment).
Length = 437
Score = 188 bits (477), Expect = 2e-46
Identities = 112/253 (44%), Positives = 150/253 (59%), Gaps = 7/253 (2%)
Frame = +2
Query: 422 LLNLLMKINAKLGGLNSLLQVEASSSIPHVSQVPTIILGMDVSHGHPGQDR-PSVAAVVS 598
L NL +KIN KLGG+N++L + P V Q P I LG DV+H G + PS+AAVV
Sbjct: 138 LSNLCLKINVKLGGVNNILLPQGR---PPVFQQPVIFLGADVTHPPAGDGKKPSIAAVVG 194
Query: 599 SRQWPLISRYRASVHTQSARLEMMSSLFKPRGTDDDGLIRESLIDFYTSSGKRKPEHIII 778
S +RY A+V Q R E++ L ++RE LI FY S+ + KP II
Sbjct: 195 SMD-AHPNRYCATVRVQQHRQEIIQDLA--------AMVRELLIQFYKST-RFKPTRIIF 244
Query: 779 FRDGVSESQFTQVINIELDQIIEACKFLDEKWSPKFTVIVAQKNHHTKFFQTASPD---- 946
+RDGVSE QF QV++ EL I EAC L++ + P T IV QK HHT+ F T +
Sbjct: 245 YRDGVSEGQFQQVLHHELLAIREACIKLEKDYQPGITFIVVQKRHHTRLFCTDKNERVGK 304
Query: 947 --NVLPGTVVDSKVCHPKNFDFYMCAHAGMIGTTRPTHYHVLHDEIGFSADEMQEFVHSL 1120
N+ GT VD+K+ HP FDFY+C+HAG+ GT+RP+HYHVL D+ FS+DE+Q + L
Sbjct: 305 SGNIPAGTTVDTKITHPTEFDFYLCSHAGIQGTSRPSHYHVLWDDNRFSSDELQILTYQL 364
Query: 1121 SYVYTIADRWIEI 1159
+ Y R + I
Sbjct: 365 CHTYVRCTRSVSI 377
>sw|O77503|I2C2_RABIT Eukaryotic translation initiation factor 2C 2
(eIF2C 2) (eIF-2C 2).
Length = 813
Score = 188 bits (477), Expect = 2e-46
Identities = 112/253 (44%), Positives = 150/253 (59%), Gaps = 7/253 (2%)
Frame = +2
Query: 422 LLNLLMKINAKLGGLNSLLQVEASSSIPHVSQVPTIILGMDVSHGHPGQDR-PSVAAVVS 598
L NL +KIN KLGG+N++L + P V Q P I LG DV+H G + PS+AAVV
Sbjct: 514 LSNLCLKINVKLGGVNNILLPQGR---PPVFQQPVIFLGADVTHPPAGDGKKPSIAAVVG 570
Query: 599 SRQWPLISRYRASVHTQSARLEMMSSLFKPRGTDDDGLIRESLIDFYTSSGKRKPEHIII 778
S +RY A+V Q R E++ L ++RE LI FY S+ + KP II
Sbjct: 571 SMD-AHPNRYCATVRVQQHRQEIIQDLA--------AMVRELLIQFYKST-RFKPTRIIF 620
Query: 779 FRDGVSESQFTQVINIELDQIIEACKFLDEKWSPKFTVIVAQKNHHTKFFQTASPD---- 946
+RDGVSE QF QV++ EL I EAC L++ + P T IV QK HHT+ F T +
Sbjct: 621 YRDGVSEGQFQQVLHHELLAIREACIKLEKDYQPGITFIVVQKRHHTRLFCTDKNERVGK 680
Query: 947 --NVLPGTVVDSKVCHPKNFDFYMCAHAGMIGTTRPTHYHVLHDEIGFSADEMQEFVHSL 1120
N+ GT VD+K+ HP FDFY+C+HAG+ GT+RP+HYHVL D+ FS+DE+Q + L
Sbjct: 681 SGNIPAGTTVDTKITHPTEFDFYLCSHAGIQGTSRPSHYHVLWDDNRFSSDELQILTYQL 740
Query: 1121 SYVYTIADRWIEI 1159
+ Y R + I
Sbjct: 741 CHTYVRCTRSVSI 753
Database: /db/trembl-ebi/tmp/swall
Posted date: Oct 24, 2003 8:22 PM
Number of letters in database: 411,996,502
Number of sequences in database: 1,288,562
Lambda K H
0.318 0.135 0.401
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 833,587,087
Number of Sequences: 1288562
Number of extensions: 16681778
Number of successful extensions: 35928
Number of sequences better than 10.0: 30
Number of HSP's better than 10.0 without gapping: 34326
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 35638
length of database: 411,996,502
effective HSP length: 126
effective length of database: 249,637,690
effective search space used: 77637321590
frameshift window, decay const: 50, 0.1
T: 12
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)

