BLASTX 2.2.6 [Apr-09-2003]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= 131537.2.170
(744 letters)
Database: /db/trembl-ebi/tmp/swall
1,288,562 sequences; 411,996,502 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
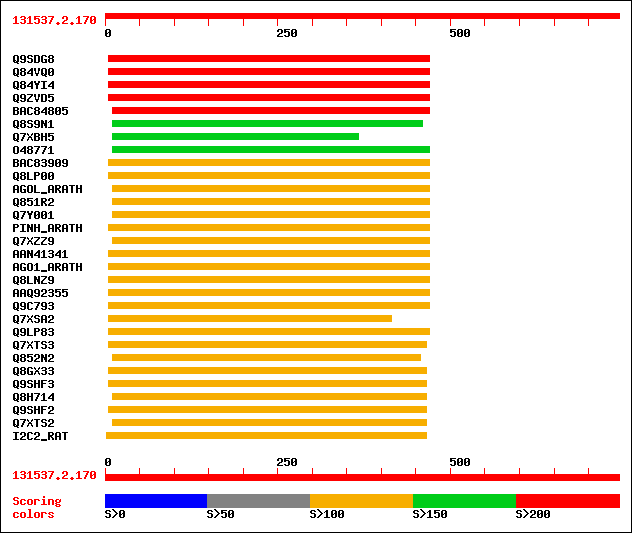
sptr|Q9SDG8|Q9SDG8 ESTs AU068544(C30430). 269 3e-71
sptr|Q84VQ0|Q84VQ0 PAZ (Piwi Argonaut and Zwille) family. 250 2e-65
sptr|Q84YI4|Q84YI4 ARGONAUTE9 protein. 248 6e-65
sptr|Q9ZVD5|Q9ZVD5 Argonaute (AGO1)-like protein (Putative post-... 242 4e-63
sptrnew|BAC84805|BAC84805 Putative ARGONAUTE9 protein. 216 2e-55
sptr|Q8S9N1|Q8S9N1 Zwille/pinhead-like protein (Fragment). 191 1e-47
sptr|Q7XBH5|Q7XBH5 Zwille pinhead-like protein. 181 1e-44
sptr|O48771|O48771 Argonaute (AGO1)-like protein. 175 5e-43
sptrnew|BAC83909|BAC83909 Putative leaf development protein Argo... 149 3e-35
sptr|Q8LP00|Q8LP00 ZLL/PNH homologous protein. 149 5e-35
sw|Q9SJK3|AGOL_ARATH Argonaute-like protein At2g27880. 147 1e-34
sptr|Q851R2|Q851R2 Putative argonaute protein. 147 1e-34
sptr|Q7Y001|Q7Y001 Putative leaf development and shoot apical me... 146 3e-34
sw|Q9XGW1|PINH_ARATH PINHEAD protein (ZWILLE protein). 145 4e-34
sptr|Q7XZZ9|Q7XZZ9 Putative leaf development and shoot apical me... 145 4e-34
sptrnew|AAN41341|AAN41341 Putative leaf development protein Argo... 143 2e-33
sw|O04379|AGO1_ARATH Argonaute protein. 143 2e-33
sptr|Q8LNZ9|Q8LNZ9 AGO1 homologous protein. 142 5e-33
sptrnew|AAQ92355|AAQ92355 ZIPPY. 140 2e-32
sptr|Q9C793|Q9C793 Pinhead-like protein. 140 2e-32
sptr|Q7XSA2|Q7XSA2 OSJNBa0005N02.3 protein. 139 3e-32
sptr|Q9LP83|Q9LP83 T1N15.2. 138 9e-32
sptr|Q7XTS3|Q7XTS3 OSJNBa0008M17.12 protein. 129 3e-29
sptr|Q852N2|Q852N2 Putative argonaute protein. 127 1e-28
sptr|Q8GX33|Q8GX33 Hypothetical protein (At1g31280). 119 5e-26
sptr|Q9SHF3|Q9SHF3 T19E23.7. 119 5e-26
sptr|Q8H714|Q8H714 Argonaute-like protein. 118 7e-26
sptr|Q9SHF2|Q9SHF2 T19E23.8. 118 7e-26
sptr|Q7XTS2|Q7XTS2 OSJNBa0008M17.13 protein. 117 2e-25
sw|Q9QZ81|I2C2_RAT Eukaryotic translation initiation factor 2C 2... 117 2e-25
>sptr|Q9SDG8|Q9SDG8 ESTs AU068544(C30430).
Length = 904
Score = 269 bits (687), Expect = 3e-71
Identities = 127/155 (81%), Positives = 141/155 (90%)
Frame = +2
Query: 5 WSPKFTVIVAQKNXHTKFFQTASPDNVLPGTVVDSKVCHPKNFDFYMCAHAGMIGTTRPT 184
WSPKFT+IVAQKN HTKFF S +NV PGTVVD+ VCHP+N DFYMCAHAGMIGTTRPT
Sbjct: 751 WSPKFTLIVAQKNHHTKFFVPGSQNNVPPGTVVDNAVCHPRNNDFYMCAHAGMIGTTRPT 810
Query: 185 HYHVLHDEIGFNADEMQEFVHSLSYVYQRSTTAISVVAPVCYAHLAAAQVSTFLRLEEMS 364
HYH+LHDEIGF+AD++QE VHSLSYVYQRSTTAISVVAP+CYAHLAAAQVS F++ +EMS
Sbjct: 811 HYHILHDEIGFSADDLQELVHSLSYVYQRSTTAISVVAPICYAHLAAAQVSQFIKFDEMS 870
Query: 365 DASSSQGGGHTSAGSAPVPELPRLHDKVRSSMFFC 469
+ SSS GGHTSAGSAPVPELPRLH+KVRSSMFFC
Sbjct: 871 ETSSSH-GGHTSAGSAPVPELPRLHNKVRSSMFFC 904
>sptr|Q84VQ0|Q84VQ0 PAZ (Piwi Argonaut and Zwille) family.
Length = 892
Score = 250 bits (638), Expect = 2e-65
Identities = 114/155 (73%), Positives = 135/155 (87%)
Frame = +2
Query: 5 WSPKFTVIVAQKNXHTKFFQTASPDNVLPGTVVDSKVCHPKNFDFYMCAHAGMIGTTRPT 184
W PKFTVIVAQKN HTKFFQ+ PDNV PGT++DS++CHP+NFDFY+CAHAGMIGTTRPT
Sbjct: 739 WHPKFTVIVAQKNHHTKFFQSRGPDNVPPGTIIDSQICHPRNFDFYLCAHAGMIGTTRPT 798
Query: 185 HYHVLHDEIGFNADEMQEFVHSLSYVYQRSTTAISVVAPVCYAHLAAAQVSTFLRLEEMS 364
HYHVL+DEIGF D++QE VHSLSYVYQRSTTAISVVAPVCYAHLAAAQ+ T ++ EE+S
Sbjct: 799 HYHVLYDEIGFATDDLQELVHSLSYVYQRSTTAISVVAPVCYAHLAAAQMGTVMKYEELS 858
Query: 365 DASSSQGGGHTSAGSAPVPELPRLHDKVRSSMFFC 469
+ SSS GG T+ G+ PVP +P+LH+ V +SMFFC
Sbjct: 859 ETSSSH-GGITTPGAVPVPPMPQLHNNVSTSMFFC 892
>sptr|Q84YI4|Q84YI4 ARGONAUTE9 protein.
Length = 896
Score = 248 bits (633), Expect = 6e-65
Identities = 113/155 (72%), Positives = 135/155 (87%)
Frame = +2
Query: 5 WSPKFTVIVAQKNXHTKFFQTASPDNVLPGTVVDSKVCHPKNFDFYMCAHAGMIGTTRPT 184
W PKFTVIVAQKN HTKFFQ+ PDNV PGT++DS++CHP+NFDFY+CAHAGMIGTTRPT
Sbjct: 743 WHPKFTVIVAQKNHHTKFFQSRGPDNVPPGTIIDSQICHPRNFDFYLCAHAGMIGTTRPT 802
Query: 185 HYHVLHDEIGFNADEMQEFVHSLSYVYQRSTTAISVVAPVCYAHLAAAQVSTFLRLEEMS 364
HYHVL+DEIGF D++QE VHSLS+VYQRSTTAISVVAPVCYAHLAAAQ+ T ++ EE+S
Sbjct: 803 HYHVLYDEIGFATDDLQELVHSLSHVYQRSTTAISVVAPVCYAHLAAAQMGTVMKYEELS 862
Query: 365 DASSSQGGGHTSAGSAPVPELPRLHDKVRSSMFFC 469
+ SSS GG T+ G+ PVP +P+LH+ V +SMFFC
Sbjct: 863 ETSSSH-GGITTPGTVPVPPMPQLHNNVSTSMFFC 896
>sptr|Q9ZVD5|Q9ZVD5 Argonaute (AGO1)-like protein (Putative
post-transcriptional gene silencing protein).
Length = 924
Score = 242 bits (617), Expect = 4e-63
Identities = 111/155 (71%), Positives = 133/155 (85%)
Frame = +2
Query: 5 WSPKFTVIVAQKNXHTKFFQTASPDNVLPGTVVDSKVCHPKNFDFYMCAHAGMIGTTRPT 184
W+PKF ++VAQKN HTKFFQ SP+NV PGT++D+K+CHPKN DFY+CAHAGMIGTTRPT
Sbjct: 771 WNPKFLLLVAQKNHHTKFFQPTSPENVPPGTIIDNKICHPKNNDFYLCAHAGMIGTTRPT 830
Query: 185 HYHVLHDEIGFNADEMQEFVHSLSYVYQRSTTAISVVAPVCYAHLAAAQVSTFLRLEEMS 364
HYHVL+DEIGF+ADE+QE VHSLSYVYQRST+AISVVAP+CYAHLAAAQ+ TF++ E+ S
Sbjct: 831 HYHVLYDEIGFSADELQELVHSLSYVYQRSTSAISVVAPICYAHLAAAQLGTFMKFEDQS 890
Query: 365 DASSSQGGGHTSAGSAPVPELPRLHDKVRSSMFFC 469
+ SSS GG T+ G V +LPRL D V +SMFFC
Sbjct: 891 ETSSSH-GGITAPGPISVAQLPRLKDNVANSMFFC 924
>sptrnew|BAC84805|BAC84805 Putative ARGONAUTE9 protein.
Length = 889
Score = 216 bits (551), Expect = 2e-55
Identities = 107/153 (69%), Positives = 124/153 (81%)
Frame = +2
Query: 11 PKFTVIVAQKNXHTKFFQTASPDNVLPGTVVDSKVCHPKNFDFYMCAHAGMIGTTRPTHY 190
PKFTVI+AQKN HTK FQ +PDNV PGTVVDS + HP+ +DFYM AHAG IGT+RPTHY
Sbjct: 738 PKFTVIIAQKNHHTKLFQENTPDNVPPGTVVDSGIVHPRQYDFYMYAHAGPIGTSRPTHY 797
Query: 191 HVLHDEIGFNADEMQEFVHSLSYVYQRSTTAISVVAPVCYAHLAAAQVSTFLRLEEMSDA 370
HVL DEIGF D++Q+ V SLSYVYQRSTTAISVVAP+CYAHLAAAQ+ F++ EE ++
Sbjct: 798 HVLLDEIGFLPDDVQKLVLSLSYVYQRSTTAISVVAPICYAHLAAAQMGQFMKFEEFAET 857
Query: 371 SSSQGGGHTSAGSAPVPELPRLHDKVRSSMFFC 469
SS GG +S+G A VPELPRLH V SSMFFC
Sbjct: 858 SSGSGGVPSSSG-AVVPELPRLHADVCSSMFFC 889
>sptr|Q8S9N1|Q8S9N1 Zwille/pinhead-like protein (Fragment).
Length = 193
Score = 191 bits (484), Expect = 1e-47
Identities = 97/150 (64%), Positives = 112/150 (74%)
Frame = +2
Query: 11 PKFTVIVAQKNXHTKFFQTASPDNVLPGTVVDSKVCHPKNFDFYMCAHAGMIGTTRPTHY 190
PKFTVI+AQKN HTK FQ +PDNV PGTVVDS + HP+ +DFYM AHAG IGT+RPTHY
Sbjct: 40 PKFTVIIAQKNHHTKLFQENTPDNVPPGTVVDSGIVHPRQYDFYMYAHAGPIGTSRPTHY 99
Query: 191 HVLHDEIGFNADEMQEFVHSLSYVYQRSTTAISVVAPVCYAHLAAAQVSTFLRLEEMSDA 370
HVL DEIGF D++Q+ V SLSYVYQRSTTAISVVAP+CYAHLAAAQ+ F++ EE ++
Sbjct: 100 HVLLDEIGFLPDDVQKLVLSLSYVYQRSTTAISVVAPICYAHLAAAQMGQFMKFEEFAEL 159
Query: 371 SSSQGGGHTSAGSAPVPELPRLHDKVRSSM 460
S PELPRLH V SSM
Sbjct: 160 RLEVVVSFVIRSSG--PELPRLHADVCSSM 187
>sptr|Q7XBH5|Q7XBH5 Zwille pinhead-like protein.
Length = 361
Score = 181 bits (458), Expect = 1e-44
Identities = 85/119 (71%), Positives = 100/119 (84%)
Frame = +2
Query: 11 PKFTVIVAQKNXHTKFFQTASPDNVLPGTVVDSKVCHPKNFDFYMCAHAGMIGTTRPTHY 190
PKFTVI+AQKN HTK FQ +PDNV PGTVVDS + HP+ +DFYM AHAG IGT+RPTHY
Sbjct: 212 PKFTVIIAQKNHHTKLFQENTPDNVPPGTVVDSGIVHPRQYDFYMYAHAGPIGTSRPTHY 271
Query: 191 HVLHDEIGFNADEMQEFVHSLSYVYQRSTTAISVVAPVCYAHLAAAQVSTFLRLEEMSD 367
HVL DEIGF D++Q+ V SLSYVYQRSTTAISVVAP+CYAHLAAAQ+ F++ EE ++
Sbjct: 272 HVLLDEIGFLPDDVQKLVLSLSYVYQRSTTAISVVAPICYAHLAAAQMGQFMKFEEFAE 330
>sptr|O48771|O48771 Argonaute (AGO1)-like protein.
Length = 887
Score = 175 bits (444), Expect = 5e-43
Identities = 90/161 (55%), Positives = 106/161 (65%), Gaps = 8/161 (4%)
Frame = +2
Query: 11 PKFTVIVAQKNXHTKFFQTASPDNVLPGTVVDSKVCHPKNFDFYMCAHAGMIGTTRPTHY 190
PKFTVIVAQKN HTK FQ P+NV GTVVD+K+ HP N+DFYMCAHAG IGT+RP HY
Sbjct: 740 PKFTVIVAQKNHHTKLFQAKGPENVPAGTVVDTKIVHPTNYDFYMCAHAGKIGTSRPAHY 799
Query: 191 HVLHDEIGFNADEMQEFVHSLSYVYQRSTTAIS--------VVAPVCYAHLAAAQVSTFL 346
HVL DEIGF+ D++Q +HSLSY S +S VAPV YAHLAAAQV+ F
Sbjct: 800 HVLLDEIGFSPDDLQNLIHSLSYKLLNSIFNVSSLLCVFVLSVAPVRYAHLAAAQVAQFT 859
Query: 347 RLEEMSDASSSQGGGHTSAGSAPVPELPRLHDKVRSSMFFC 469
+ E +S+ VPELPRLH+ V +MFFC
Sbjct: 860 KFEGISE-------------DGKVPELPRLHENVEGNMFFC 887
>sptrnew|BAC83909|BAC83909 Putative leaf development protein
Argonaute.
Length = 1052
Score = 149 bits (377), Expect = 3e-35
Identities = 76/166 (45%), Positives = 100/166 (60%), Gaps = 11/166 (6%)
Frame = +2
Query: 5 WSPKFTVIVAQKNXHTKFFQ--------TASPDNVLPGTVVDSKVCHPKNFDFYMCAHAG 160
+ P T +V QK HT+ F T NV PGTVVD+ +CHP FDFY+C+HAG
Sbjct: 887 YRPLVTFVVVQKRHHTRLFPEVHGRQDLTDRSGNVRPGTVVDTNICHPSEFDFYLCSHAG 946
Query: 161 MIGTTRPTHYHVLHDEIGFNADEMQEFVHSLSYVYQRSTTAISVVAPVCYAHLAAAQVST 340
+ GT+RPTHYHVLHDE F+AD++Q ++L Y Y R T ++SVV P YAHLAA +
Sbjct: 947 IQGTSRPTHYHVLHDENRFSADQLQMLTYNLCYTYARCTRSVSVVPPAYYAHLAAFRARY 1006
Query: 341 FLRLEEMSDASS-SQGGGHTSAGSAP--VPELPRLHDKVRSSMFFC 469
+ M ASS GG +AG P V LP++ + V+ MF+C
Sbjct: 1007 YDEPPAMDGASSVGSGGNQAAAGGQPPAVRRLPQIKENVKDVMFYC 1052
>sptr|Q8LP00|Q8LP00 ZLL/PNH homologous protein.
Length = 978
Score = 149 bits (375), Expect = 5e-35
Identities = 73/163 (44%), Positives = 97/163 (59%), Gaps = 8/163 (4%)
Frame = +2
Query: 5 WSPKFTVIVAQKNXHTKFFQTASPD--------NVLPGTVVDSKVCHPKNFDFYMCAHAG 160
+ P T +V QK HT+ F D N+LPGTVVDSK+CHP FDFY+C+HAG
Sbjct: 818 YQPPVTFVVVQKRHHTRLFANNHKDRSSTDKSGNILPGTVVDSKICHPSEFDFYLCSHAG 877
Query: 161 MIGTTRPTHYHVLHDEIGFNADEMQEFVHSLSYVYQRSTTAISVVAPVCYAHLAAAQVST 340
+ GT+RP HYHVL DE F ADEMQ ++L Y Y R T ++SVV P YAHLAA +
Sbjct: 878 IQGTSRPAHYHVLWDENNFTADEMQTLTNNLCYTYARCTRSVSVVPPAYYAHLAAFRARF 937
Query: 341 FLRLEEMSDASSSQGGGHTSAGSAPVPELPRLHDKVRSSMFFC 469
++ E + ++S+ T V LP + +KV+ MF+C
Sbjct: 938 YMEPEMSENQTTSKSS--TGTNGTSVKPLPAVKEKVKRVMFYC 978
>sw|Q9SJK3|AGOL_ARATH Argonaute-like protein At2g27880.
Length = 997
Score = 147 bits (371), Expect = 1e-34
Identities = 70/161 (43%), Positives = 98/161 (60%), Gaps = 8/161 (4%)
Frame = +2
Query: 11 PKFTVIVAQKNXHTKFF--------QTASPDNVLPGTVVDSKVCHPKNFDFYMCAHAGMI 166
P+ T ++ QK HT+ F T N+ PGTVVD+K+CHP FDFY+ +HAG+
Sbjct: 838 PRVTFVIVQKRHHTRLFPEQHGNRDMTDKSGNIQPGTVVDTKICHPNEFDFYLNSHAGIQ 897
Query: 167 GTTRPTHYHVLHDEIGFNADEMQEFVHSLSYVYQRSTTAISVVAPVCYAHLAAAQVSTFL 346
GT+RP HYHVL DE GF AD++Q ++L Y Y R T ++S+V P YAHLAA + ++
Sbjct: 898 GTSRPAHYHVLLDENGFTADQLQMLTNNLCYTYARCTKSVSIVPPAYYAHLAAFRARYYM 957
Query: 347 RLEEMSDASSSQGGGHTSAGSAPVPELPRLHDKVRSSMFFC 469
EMSD SS+ T+ + +LP + D V+ MF+C
Sbjct: 958 E-SEMSDGGSSRSRSSTTGVGQVISQLPAIKDNVKEVMFYC 997
>sptr|Q851R2|Q851R2 Putative argonaute protein.
Length = 1058
Score = 147 bits (371), Expect = 1e-34
Identities = 72/163 (44%), Positives = 96/163 (58%), Gaps = 10/163 (6%)
Frame = +2
Query: 11 PKFTVIVAQKNXHTKFF--------QTASPDNVLPGTVVDSKVCHPKNFDFYMCAHAGMI 166
P T +V QK HT+ F T N+LPGTVVD ++CHP FDFY+C+HAG+
Sbjct: 896 PPVTFVVVQKRHHTRLFPEVHGRRDMTDKSGNILPGTVVDRQICHPTEFDFYLCSHAGIQ 955
Query: 167 GTTRPTHYHVLHDEIGFNADEMQEFVHSLSYVYQRSTTAISVVAPVCYAHLAAAQVSTFL 346
GT+RPTHYHVL+DE F AD +Q ++L Y Y R T A+SVV P YAHLAA + ++
Sbjct: 956 GTSRPTHYHVLYDENHFTADALQSLTNNLCYTYARCTRAVSVVPPAYYAHLAAFRARYYV 1015
Query: 347 RLEEMSDASSSQGGGHTSAGSAPVP--ELPRLHDKVRSSMFFC 469
E S+ G A PV +LP++ + V+ MF+C
Sbjct: 1016 EGESSDGGSTPGSSGQAVAREGPVEVRQLPKIKENVKDVMFYC 1058
>sptr|Q7Y001|Q7Y001 Putative leaf development and shoot apical
meristem regulating protein.
Length = 1055
Score = 146 bits (368), Expect = 3e-34
Identities = 71/159 (44%), Positives = 95/159 (59%), Gaps = 6/159 (3%)
Frame = +2
Query: 11 PKFTVIVAQKNXHTKFFQTASPD------NVLPGTVVDSKVCHPKNFDFYMCAHAGMIGT 172
P T +V QK HT+ F D N+LPGTVVD+ +CHP FDFY+C+H+G+ GT
Sbjct: 900 PPVTFVVVQKRHHTRLFPENRRDMMDRSGNILPGTVVDTMICHPSEFDFYLCSHSGIKGT 959
Query: 173 TRPTHYHVLHDEIGFNADEMQEFVHSLSYVYQRSTTAISVVAPVCYAHLAAAQVSTFLRL 352
+RPTHYHVL DE GF AD +Q ++LSY Y R T A+S+V P YAHL A + ++
Sbjct: 960 SRPTHYHVLLDENGFKADTLQTLTYNLSYTYARCTRAVSIVPPAYYAHLGAFRARYYMED 1019
Query: 353 EEMSDASSSQGGGHTSAGSAPVPELPRLHDKVRSSMFFC 469
E SSS T + P+PE + + V+ MF+C
Sbjct: 1020 EHSDQGSSSSVTTRTDRSTKPLPE---IKENVKRFMFYC 1055
>sw|Q9XGW1|PINH_ARATH PINHEAD protein (ZWILLE protein).
Length = 988
Score = 145 bits (367), Expect = 4e-34
Identities = 74/165 (44%), Positives = 96/165 (58%), Gaps = 10/165 (6%)
Frame = +2
Query: 5 WSPKFTVIVAQKNXHTKFFQTASPD--------NVLPGTVVDSKVCHPKNFDFYMCAHAG 160
+ P T IV QK HT+ F D N+LPGTVVD+K+CHP FDFY+C+HAG
Sbjct: 824 YQPPVTFIVVQKRHHTRLFANNHRDKNSTDRSGNILPGTVVDTKICHPTEFDFYLCSHAG 883
Query: 161 MIGTTRPTHYHVLHDEIGFNADEMQEFVHSLSYVYQRSTTAISVVAPVCYAHLAAAQVST 340
+ GT+RP HYHVL DE F AD +Q ++L Y Y R T ++S+V P YAHLAA +
Sbjct: 884 IQGTSRPAHYHVLWDENNFTADGIQSLTNNLCYTYARCTRSVSIVPPAYYAHLAAFRARF 943
Query: 341 FLRLEEMSDASS--SQGGGHTSAGSAPVPELPRLHDKVRSSMFFC 469
+L E M D S + T+ G V LP L + V+ MF+C
Sbjct: 944 YLEPEIMQDNGSPGKKNTKTTTVGDVGVKPLPALKENVKRVMFYC 988
>sptr|Q7XZZ9|Q7XZZ9 Putative leaf development and shoot apical
meristem regulating protein.
Length = 895
Score = 145 bits (367), Expect = 4e-34
Identities = 70/165 (42%), Positives = 99/165 (60%), Gaps = 12/165 (7%)
Frame = +2
Query: 11 PKFTVIVAQKNXHTKFF--------QTASPDNVLPGTVVDSKVCHPKNFDFYMCAHAGMI 166
P T +V QK HT+ F Q N+LPGTVVD+K+CHP FDFY+C+H+G+
Sbjct: 731 PPVTFVVVQKRHHTRLFPEDHHARDQMDRSRNILPGTVVDTKICHPSEFDFYLCSHSGIQ 790
Query: 167 GTTRPTHYHVLHDEIGFNADEMQEFVHSLSYVYQRSTTAISVVAPVCYAHLAAAQVSTFL 346
GT+ PTHY+VL DE F+AD +Q + L Y Y R T ++S+V PV YAHLAA++ +L
Sbjct: 791 GTSHPTHYYVLFDENNFSADALQTLTYHLCYTYARCTRSVSIVPPVYYAHLAASRARHYL 850
Query: 347 RLEEMSD----ASSSQGGGHTSAGSAPVPELPRLHDKVRSSMFFC 469
+ D ++S+ GG + PV LP + + V+ MF+C
Sbjct: 851 EEGSLPDHGSSSASAAGGSRRNDRGVPVKPLPEIKENVKQFMFYC 895
>sptrnew|AAN41341|AAN41341 Putative leaf development protein
Argonaute.
Length = 1048
Score = 143 bits (361), Expect = 2e-33
Identities = 74/174 (42%), Positives = 99/174 (56%), Gaps = 19/174 (10%)
Frame = +2
Query: 5 WSPKFTVIVAQKNXHTKFFQTASPD--------NVLPGTVVDSKVCHPKNFDFYMCAHAG 160
+ P T +V QK HT+ F D N+LPGTVVDSK+CHP FDFY+C+HAG
Sbjct: 875 YQPPVTFVVVQKRHHTRLFAQNHNDRHSVDRSGNILPGTVVDSKICHPTEFDFYLCSHAG 934
Query: 161 MIGTTRPTHYHVLHDEIGFNADEMQEFVHSLSYVYQRSTTAISVVAPVCYAHLAAAQVST 340
+ GT+RP HYHVL DE F AD +Q ++L Y Y R T ++S+V P YAHLAA +
Sbjct: 935 IQGTSRPAHYHVLWDENNFTADGLQSLTNNLCYTYARCTRSVSIVPPAYYAHLAAFRARF 994
Query: 341 FLRLE-----EMSDASSSQGGGHTSAG------SAPVPELPRLHDKVRSSMFFC 469
++ E M+ S ++GGG +A V LP L + V+ MF+C
Sbjct: 995 YMEPETSDSGSMASGSMARGGGMAGRSTRGPNVNAAVRPLPALKENVKRVMFYC 1048
>sw|O04379|AGO1_ARATH Argonaute protein.
Length = 1048
Score = 143 bits (361), Expect = 2e-33
Identities = 74/174 (42%), Positives = 99/174 (56%), Gaps = 19/174 (10%)
Frame = +2
Query: 5 WSPKFTVIVAQKNXHTKFFQTASPD--------NVLPGTVVDSKVCHPKNFDFYMCAHAG 160
+ P T +V QK HT+ F D N+LPGTVVDSK+CHP FDFY+C+HAG
Sbjct: 875 YQPPVTFVVVQKRHHTRLFAQNHNDRHSVDRSGNILPGTVVDSKICHPTEFDFYLCSHAG 934
Query: 161 MIGTTRPTHYHVLHDEIGFNADEMQEFVHSLSYVYQRSTTAISVVAPVCYAHLAAAQVST 340
+ GT+RP HYHVL DE F AD +Q ++L Y Y R T ++S+V P YAHLAA +
Sbjct: 935 IQGTSRPAHYHVLWDENNFTADGLQSLTNNLCYTYARCTRSVSIVPPAYYAHLAAFRARF 994
Query: 341 FLRLE-----EMSDASSSQGGGHTSAG------SAPVPELPRLHDKVRSSMFFC 469
++ E M+ S ++GGG +A V LP L + V+ MF+C
Sbjct: 995 YMEPETSDSGSMASGSMARGGGMAGRSTRGPNVNAAVRPLPALKENVKRVMFYC 1048
>sptr|Q8LNZ9|Q8LNZ9 AGO1 homologous protein.
Length = 909
Score = 142 bits (358), Expect = 5e-33
Identities = 76/175 (43%), Positives = 98/175 (56%), Gaps = 20/175 (11%)
Frame = +2
Query: 5 WSPKFTVIVAQKNXHTKFFQTASPD--------NVLPGTVVDSKVCHPKNFDFYMCAHAG 160
+ PK T IV QK HT+ F D N+LPGTVVDSK+CHP FDFY+C+HAG
Sbjct: 735 YQPKVTFIVVQKRHHTRLFAHNHNDQNSVDRSGNILPGTVVDSKICHPTEFDFYLCSHAG 794
Query: 161 MIGTTRPTHYHVLHDEIGFNADEMQEFVHSLSYVYQRSTTAISVVAPVCYAHLAAAQVST 340
+ GT+RP HYHVL +E F AD +Q ++L Y Y R T ++S+V P YAHLAA +
Sbjct: 795 IKGTSRPAHYHVLWNENNFTADALQILTNNLCYTYARCTRSVSIVPPAYYAHLAAFRARF 854
Query: 341 FLRLEEMSDASSSQG--------GGHTS----AGSAPVPELPRLHDKVRSSMFFC 469
++ + +S G G TS G A V LP L D V+ MF+C
Sbjct: 855 YMEPDTSDSSSVVSGPGVRGPLSGSSTSRTRAPGGAAVKPLPALKDSVKRVMFYC 909
>sptrnew|AAQ92355|AAQ92355 ZIPPY.
Length = 990
Score = 140 bits (353), Expect = 2e-32
Identities = 68/157 (43%), Positives = 95/157 (60%), Gaps = 2/157 (1%)
Frame = +2
Query: 5 WSPKFTVIVAQKNXHTKFFQTASPD--NVLPGTVVDSKVCHPKNFDFYMCAHAGMIGTTR 178
++P T V QK HT+ F+ PD N+ PGTVVD+ + HPK FDFY+C+H G+ GT+R
Sbjct: 835 YNPSITFAVVQKRHHTRLFR-CDPDHENIPPGTVVDTVITHPKEFDFYLCSHLGVKGTSR 893
Query: 179 PTHYHVLHDEIGFNADEMQEFVHSLSYVYQRSTTAISVVAPVCYAHLAAAQVSTFLRLEE 358
PTHYH+L DE F +DE+Q V++L Y + R T IS+V P YAHLAA + ++
Sbjct: 894 PTHYHILWDENEFTSDELQRLVYNLCYTFVRCTKPISIVPPAYYAHLAAYRGRLYIERSS 953
Query: 359 MSDASSSQGGGHTSAGSAPVPELPRLHDKVRSSMFFC 469
S+ S + G LP+L D V++ MF+C
Sbjct: 954 ESNGGSMNPSSVSRVGPPKTIPLPKLSDNVKNLMFYC 990
>sptr|Q9C793|Q9C793 Pinhead-like protein.
Length = 990
Score = 140 bits (353), Expect = 2e-32
Identities = 68/157 (43%), Positives = 95/157 (60%), Gaps = 2/157 (1%)
Frame = +2
Query: 5 WSPKFTVIVAQKNXHTKFFQTASPD--NVLPGTVVDSKVCHPKNFDFYMCAHAGMIGTTR 178
++P T V QK HT+ F+ PD N+ PGTVVD+ + HPK FDFY+C+H G+ GT+R
Sbjct: 835 YNPSITFAVVQKRHHTRLFR-CDPDHENIPPGTVVDTVITHPKEFDFYLCSHLGVKGTSR 893
Query: 179 PTHYHVLHDEIGFNADEMQEFVHSLSYVYQRSTTAISVVAPVCYAHLAAAQVSTFLRLEE 358
PTHYH+L DE F +DE+Q V++L Y + R T IS+V P YAHLAA + ++
Sbjct: 894 PTHYHILWDENEFTSDELQRLVYNLCYTFVRCTKPISIVPPAYYAHLAAYRGRLYIERSS 953
Query: 359 MSDASSSQGGGHTSAGSAPVPELPRLHDKVRSSMFFC 469
S+ S + G LP+L D V++ MF+C
Sbjct: 954 ESNGGSMNPSSVSRVGPPKTIPLPKLSDNVKNLMFYC 990
>sptr|Q7XSA2|Q7XSA2 OSJNBa0005N02.3 protein.
Length = 1254
Score = 139 bits (351), Expect = 3e-32
Identities = 69/145 (47%), Positives = 88/145 (60%), Gaps = 8/145 (5%)
Frame = +2
Query: 5 WSPKFTVIVAQKNXHTKFFQTASPD--------NVLPGTVVDSKVCHPKNFDFYMCAHAG 160
+ P T +V QK HT+ F D N+LPGTVVDSK+CHP FDFY+C+HAG
Sbjct: 1031 YQPPVTFVVVQKRHHTRLFANNHNDQRTVDRSGNILPGTVVDSKICHPTEFDFYLCSHAG 1090
Query: 161 MIGTTRPTHYHVLHDEIGFNADEMQEFVHSLSYVYQRSTTAISVVAPVCYAHLAAAQVST 340
+ GT+RP HYHVL DE F ADE+Q ++L Y Y R T ++S+V P YAHLAA +
Sbjct: 1091 IQGTSRPAHYHVLWDENKFTADELQTLTNNLCYTYARCTRSVSIVPPAYYAHLAAFRARF 1150
Query: 341 FLRLEEMSDASSSQGGGHTSAGSAP 415
++ E SD+ S G TS G P
Sbjct: 1151 YME-PETSDSGSMASGAATSRGLPP 1174
>sptr|Q9LP83|Q9LP83 T1N15.2.
Length = 1123
Score = 138 bits (347), Expect = 9e-32
Identities = 74/177 (41%), Positives = 99/177 (55%), Gaps = 22/177 (12%)
Frame = +2
Query: 5 WSPKFTVIVAQKNXHTKFFQTASPD--------NVLPG---TVVDSKVCHPKNFDFYMCA 151
+ P T +V QK HT+ F D N+LPG TVVDSK+CHP FDFY+C+
Sbjct: 947 YQPPVTFVVVQKRHHTRLFAQNHNDRHSVDRSGNILPGETSTVVDSKICHPTEFDFYLCS 1006
Query: 152 HAGMIGTTRPTHYHVLHDEIGFNADEMQEFVHSLSYVYQRSTTAISVVAPVCYAHLAAAQ 331
HAG+ GT+RP HYHVL DE F AD +Q ++L Y Y R T ++S+V P YAHLAA +
Sbjct: 1007 HAGIQGTSRPAHYHVLWDENNFTADGLQSLTNNLCYTYARCTRSVSIVPPAYYAHLAAFR 1066
Query: 332 VSTFLRLE-----EMSDASSSQGGGHTSAG------SAPVPELPRLHDKVRSSMFFC 469
++ E M+ S ++GGG +A V LP L + V+ MF+C
Sbjct: 1067 ARFYMEPETSDSGSMASGSMARGGGMAGRSTRGPNVNAAVRPLPALKENVKRVMFYC 1123
>sptr|Q7XTS3|Q7XTS3 OSJNBa0008M17.12 protein.
Length = 1034
Score = 129 bits (325), Expect = 3e-29
Identities = 69/165 (41%), Positives = 99/165 (60%), Gaps = 11/165 (6%)
Frame = +2
Query: 5 WSPKFTVIVAQKNXHTKFF------QTASPDNVLPGTVVDSKVCHPKNFDFYMCAHAGMI 166
+SP TVIVA+K HT+ F Q NVLPGTVVD+ V P +DFY+C+H G+I
Sbjct: 869 YSPTITVIVAKKRHHTRLFPKDLNQQQTKNGNVLPGTVVDTGVVDPAAYDFYLCSHNGLI 928
Query: 167 GTTRPTHYHVLHDEIGFNADEMQEFVHSLSYVYQRSTTAISVVAPVCYAHLAAAQVSTFL 346
GT+RPTHY+ L DE GF +D++Q+ V++L +V+ R T +S+ PV YA LAA + +
Sbjct: 929 GTSRPTHYYSLLDEHGFASDDLQKLVYNLCFVFARCTKPVSLATPVYYADLAAYRGRLYY 988
Query: 347 RLEEM-----SDASSSQGGGHTSAGSAPVPELPRLHDKVRSSMFF 466
M S A+S+ + AG++ P LH+ + +MFF
Sbjct: 989 EGMMMSQPPPSSAASASSASSSGAGASDFRSFPALHEDLVDNMFF 1033
>sptr|Q852N2|Q852N2 Putative argonaute protein.
Length = 1192
Score = 127 bits (320), Expect = 1e-28
Identities = 69/158 (43%), Positives = 92/158 (58%), Gaps = 9/158 (5%)
Frame = +2
Query: 11 PKFTVIVAQKNXHTKFF--------QTASPDNVLPGTVVDSKVCHPKNFDFYMCAHAGMI 166
P T +V QK HT+ F T N+LPGTV D ++CHP F FY+C+HAG+
Sbjct: 650 PPVTFVVVQKRHHTRLFPEVHGRRDMTDKSGNILPGTVKDRQICHPTEFYFYLCSHAGIQ 709
Query: 167 GTTRPTHYHVLHDEIGFNADEMQEFVHSLSYVYQRSTTAISVVAPVCYAHLAAAQVSTFL 346
GT+RPTHYHVL+DE F ADE+Q ++L Y+Y R T A+SVV P Y+HLAA+ +
Sbjct: 710 GTSRPTHYHVLYDENHFTADELQTLTNNLCYIYARCTHAVSVVPPAYYSHLAASHAHCCI 769
Query: 347 RLEEMSDASSSQGGGHTS-AGSAPVPELPRLHDKVRSS 457
+ GH+S +GS P E HD V++S
Sbjct: 770 K-------------GHSSGSGSTPGNE----HDIVKNS 790
>sptr|Q8GX33|Q8GX33 Hypothetical protein (At1g31280).
Length = 540
Score = 119 bits (297), Expect = 5e-26
Identities = 63/168 (37%), Positives = 93/168 (55%), Gaps = 14/168 (8%)
Frame = +2
Query: 5 WSPKFTVIVAQKNXHTKFFQTASPD-----NVLPGTVVDSKVCHPKNFDFYMCAHAGMIG 169
++PK TVIVAQK T+FF + D NV GTVVD+KV HP +DFY+C+H G IG
Sbjct: 372 YNPKITVIVAQKRHQTRFFPATNNDGSDKGNVPSGTVVDTKVIHPYEYDFYLCSHHGGIG 431
Query: 170 TTRPTHYHVLHDEIGFNADEMQEFVHSLSYVYQRSTTAISVVAPVCYAHLAAAQVSTFLR 349
T++PTHY+ L DE+GF +D++Q+ + + + + R T +S+V PV YA + A + +
Sbjct: 432 TSKPTHYYTLWDELGFTSDQVQKLIFEMCFTFTRCTKPVSLVPPVYYADMVAFRGRMYHE 491
Query: 350 LEEMSDASSSQGGGHTSAGSAPV---------PELPRLHDKVRSSMFF 466
G TSA S + +LH ++ + MFF
Sbjct: 492 ASSREKNFKQPRGASTSAASLASSLSSLTIEDKAIFKLHAELENVMFF 539
>sptr|Q9SHF3|Q9SHF3 T19E23.7.
Length = 1014
Score = 119 bits (297), Expect = 5e-26
Identities = 63/168 (37%), Positives = 93/168 (55%), Gaps = 14/168 (8%)
Frame = +2
Query: 5 WSPKFTVIVAQKNXHTKFFQTASPD-----NVLPGTVVDSKVCHPKNFDFYMCAHAGMIG 169
++PK TVIVAQK T+FF + D NV GTVVD+KV HP +DFY+C+H G IG
Sbjct: 846 YNPKITVIVAQKRHQTRFFPATNNDGSDKGNVPSGTVVDTKVIHPYEYDFYLCSHHGGIG 905
Query: 170 TTRPTHYHVLHDEIGFNADEMQEFVHSLSYVYQRSTTAISVVAPVCYAHLAAAQVSTFLR 349
T++PTHY+ L DE+GF +D++Q+ + + + + R T +S+V PV YA + A + +
Sbjct: 906 TSKPTHYYTLWDELGFTSDQVQKLIFEMCFTFTRCTKPVSLVPPVYYADMVAFRGRMYHE 965
Query: 350 LEEMSDASSSQGGGHTSAGSAPV---------PELPRLHDKVRSSMFF 466
G TSA S + +LH ++ + MFF
Sbjct: 966 ASSREKNFKQPRGASTSAASLASSLSSLTIEDKAIFKLHAELENVMFF 1013
>sptr|Q8H714|Q8H714 Argonaute-like protein.
Length = 210
Score = 118 bits (296), Expect = 7e-26
Identities = 64/157 (40%), Positives = 90/157 (57%), Gaps = 5/157 (3%)
Frame = +2
Query: 11 PKFTVIVAQKNXHTKFFQTASPD-----NVLPGTVVDSKVCHPKNFDFYMCAHAGMIGTT 175
P T ++ QK +T+ F D NV GTVVD+ +CHP DFY+ +HAG+ GT+
Sbjct: 56 PPITFVIVQKRHNTRLFPDNPKDADRSGNVKAGTVVDTGICHPIENDFYLMSHAGLQGTS 115
Query: 176 RPTHYHVLHDEIGFNADEMQEFVHSLSYVYQRSTTAISVVAPVCYAHLAAAQVSTFLRLE 355
RPTHYHVL +EIGF ADE+Q + L Y + R T ++S+V Y+HL A + F L
Sbjct: 116 RPTHYHVLLNEIGFTADELQTLTYKLCYTFARCTRSVSMVPSAYYSHLVAFRARFF--LA 173
Query: 356 EMSDASSSQGGGHTSAGSAPVPELPRLHDKVRSSMFF 466
E SD +S+ G +A + LH ++S M+F
Sbjct: 174 EGSDTASTVSGFSETAPETDT-RMYDLHQAMKSEMYF 209
>sptr|Q9SHF2|Q9SHF2 T19E23.8.
Length = 1194
Score = 118 bits (296), Expect = 7e-26
Identities = 64/169 (37%), Positives = 97/169 (57%), Gaps = 15/169 (8%)
Frame = +2
Query: 5 WSPKFTVIVAQKNXHTKFFQ-TASPD-----NVLPGTVVDSKVCHPKNFDFYMCAHAGMI 166
++P+ TVIVAQK T+FF T S D NV GTVVD+ + HP +DFY+C+ G I
Sbjct: 1025 YNPQITVIVAQKRHQTRFFPATTSKDGRAKGNVPSGTVVDTTIIHPFEYDFYLCSQHGAI 1084
Query: 167 GTTRPTHYHVLHDEIGFNADEMQEFVHSLSYVYQRSTTAISVVAPVCYAHLAAAQVSTFL 346
GT++PTHY+VL DEIGFN++++Q+ + L + + R T +++V PV YA AA++ +
Sbjct: 1085 GTSKPTHYYVLSDEIGFNSNQIQKLIFDLCFTFTRCTKPVALVPPVSYADKAASRGRVYY 1144
Query: 347 RLEEMSDASSSQGGGHTSAGSAPV---------PELPRLHDKVRSSMFF 466
M S G +S+ S E+ ++H + + MFF
Sbjct: 1145 EASLMKKNSKQSRGASSSSASVASSSSSVTMEDKEIFKVHAGIENFMFF 1193
>sptr|Q7XTS2|Q7XTS2 OSJNBa0008M17.13 protein.
Length = 1109
Score = 117 bits (293), Expect = 2e-25
Identities = 73/204 (35%), Positives = 101/204 (49%), Gaps = 52/204 (25%)
Frame = +2
Query: 11 PKFTVIVAQKNXHTKFF------QTASPDNVLPGTVVDSKVCHPKNFDFYMCAHAGMIGT 172
PK TVIVA+K HT+ F + NVLPGTVVD+ V P +DFY+C+H G +GT
Sbjct: 905 PKITVIVAKKRHHTRLFPKDRNQRQTKNGNVLPGTVVDTDVVDPTAYDFYLCSHKGEVGT 964
Query: 173 TRPTHYHVLHDEIGFNADEMQEFVHSLSYVYQRSTTAISVVAPVCYAHLA---------- 322
+RPTHY+ L DE GF +D++Q+ V++L +V+ R T +S+ PV YA LA
Sbjct: 965 SRPTHYYSLLDEHGFASDDLQKLVYNLCFVFARCTKPVSLATPVYYADLAAYRGRLYYEG 1024
Query: 323 ------------------------AAQVSTFLRLEEMSDASSSQG------------GGH 394
AA + S ASSS+G
Sbjct: 1025 MMMLQPAASAASASEAMMPAAQPQAAAAAAAAASPSSSAASSSEGMTASQPQAPAAEAAS 1084
Query: 395 TSAGSAPVPELPRLHDKVRSSMFF 466
+SAG+A ELP +H + ++MFF
Sbjct: 1085 SSAGAADFRELPPMHGDLLNNMFF 1108
>sw|Q9QZ81|I2C2_RAT Eukaryotic translation initiation factor 2C 2
(eIF2C 2) (eIF-2C 2) (Golgi ER protein 95 kDa) (GERp95).
Length = 863
Score = 117 bits (292), Expect = 2e-25
Identities = 60/162 (37%), Positives = 91/162 (56%), Gaps = 7/162 (4%)
Frame = +2
Query: 2 KWSPKFTVIVAQKNXHTKFFQTASPD------NVLPGTVVDSKVCHPKNFDFYMCAHAGM 163
++ P T IV QK HT+ F T + N+ GT VD+K+ HP FDFY+C+HAG+
Sbjct: 701 EYQPGITFIVVQKRHHTRLFCTDKNERVGKSGNIPAGTTVDTKITHPTEFDFYLCSHAGI 760
Query: 164 IGTTRPTHYHVLHDEIGFNADEMQEFVHSLSYVYQRSTTAISVVAPVCYAHLAAAQVSTF 343
GT+RP+HYHVL D+ F++DE+Q + L + Y R T ++S+ AP YAHL A +
Sbjct: 761 QGTSRPSHYHVLWDDNRFSSDELQILTYQLCHTYVRCTRSVSIPAPAYYAHLVAFRARYH 820
Query: 344 LRLEEMSDASSSQGGGHTSA-GSAPVPELPRLHDKVRSSMFF 466
L +E A S G ++ + + ++H +M+F
Sbjct: 821 LVDKEHDSAEGSHTSGQSNGRDHQALAKAVQVHQDTLRTMYF 862
Database: /db/trembl-ebi/tmp/swall
Posted date: Oct 24, 2003 8:22 PM
Number of letters in database: 411,996,502
Number of sequences in database: 1,288,562
Lambda K H
0.318 0.135 0.401
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 604,043,705
Number of Sequences: 1288562
Number of extensions: 13556334
Number of successful extensions: 39805
Number of sequences better than 10.0: 30
Number of HSP's better than 10.0 without gapping: 37531
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 39734
length of database: 411,996,502
effective HSP length: 120
effective length of database: 257,369,062
effective search space used: 32685870874
frameshift window, decay const: 50, 0.1
T: 12
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)

