BLASTX 2.2.6 [Apr-09-2003]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= 129373.2.1
(1297 letters)
Database: /db/trembl-ebi/tmp/swall
1,272,877 sequences; 406,886,216 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
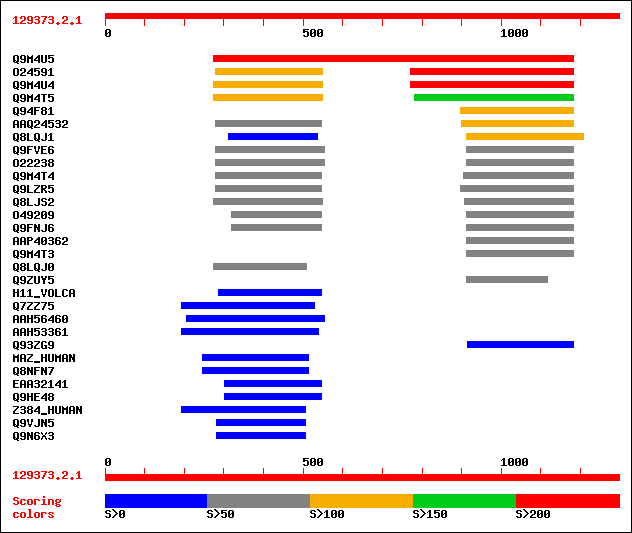
sptr|Q9M4U5|Q9M4U5 Histone deacetylase 2 isoform b. 408 e-112
sptr|O24591|O24591 Nucleolar histone deacetylase HD2-P39. 209 7e-53
sptr|Q9M4U4|Q9M4U4 Histone deacetylase 2 isoform c. 207 2e-52
sptr|Q9M4T5|Q9M4T5 Putative histone deacetylase HD2. 164 3e-39
sptr|Q94F81|Q94F81 HD2 type histone deacetylase HDA106. 124 2e-27
sptrnew|AAQ24532|AAQ24532 Histone deacetylase. 112 2e-23
sptr|Q8LQJ1|Q8LQJ1 Histone deacetylase HD2-like protein. 103 5e-21
sptr|Q9FVE6|Q9FVE6 Putative histone deacetylase. 99 1e-19
sptr|O22238|O22238 Putative histone deacetylase. 99 1e-19
sptr|Q9M4T4|Q9M4T4 Putative histone deacetylase HD2c (AT5g03740/... 98 2e-19
sptr|Q9LZR5|Q9LZR5 Histone deacetylase-like protein. 97 4e-19
sptr|Q8LJS2|Q8LJS2 Nucleolar histone deacetylase HD2-P39. 96 9e-19
sptr|O49209|O49209 Putative histone deacetylase. 93 1e-17
sptr|Q9FNJ6|Q9FNJ6 Histone deacetylase-like protein (AT5g22650/M... 93 1e-17
sptrnew|AAP40362|AAP40362 Hypothetical protein At2g27840. 78 2e-13
sptr|Q9M4T3|Q9M4T3 Putative histone deacetylase HD2d (Hypothetic... 78 2e-13
sptr|Q8LQJ0|Q8LQJ0 P0456E05.12 protein. 55 3e-06
sptr|Q9ZUY5|Q9ZUY5 Hypothetical 20.1 kDa protein. 52 2e-05
sw|Q08864|H11_VOLCA Histone H1-I. 42 0.025
sptr|Q7ZZ75|Q7ZZ75 SI:dZ44O19.5 (Novel zinc finger protein (5 do... 41 0.033
sptrnew|AAH56460|AAH56460 Hypothetical protein (Fragment). 40 0.096
sptrnew|AAH53361|AAH53361 Hypothetical protein. 40 0.096
sptr|Q93ZG9|Q93ZG9 AT4g25340/T30C3_20. 40 0.096
sw|P56270|MAZ_HUMAN Myc-associated zinc finger protein (MAZI) (P... 40 0.096
sptr|Q8NFN7|Q8NFN7 Serum amyloid A activating factor 2. 40 0.096
sptrnew|EAA32141|EAA32141 Hypothetical protein ( (AL451013) prob... 40 0.096
sptr|Q9HE48|Q9HE48 Probable nucleotide exsicion repair protein R... 40 0.096
sw|Q8TF68|Z384_HUMAN Zinc finger protein 384 (Nuclear matrix tra... 39 0.13
sptr|Q9VJN5|Q9VJN5 CG4148 protein (LD22579P). 39 0.13
sptr|Q9N6X3|Q9N6X3 L(2)35Ea protein. 39 0.13
>sptr|Q9M4U5|Q9M4U5 Histone deacetylase 2 isoform b.
Length = 303
Score = 408 bits (1048), Expect = e-112
Identities = 218/303 (71%), Positives = 218/303 (71%)
Frame = -1
Query: 1183 MEFWGLEVKPGSTVKCEPGHGFILHVSQAALGESKKSDSALMYVKVDDKKLAIGTLSIDK 1004
MEFWGLEVKPGSTVKCEPGHGFILHVSQAALGESKKSDSALMYVKVDDKKLAIGTLSIDK
Sbjct: 1 MEFWGLEVKPGSTVKCEPGHGFILHVSQAALGESKKSDSALMYVKVDDKKLAIGTLSIDK 60
Query: 1003 YPQIQFDLVFNKEFELSHTSKTTSVFFSGYKVEQPIXXXXXXXXXXXXXXELNIPVIKEN 824
YPQIQFDLVFNKEFELSHTSKTTSVFFSGYKVEQPI ELNIPVIKEN
Sbjct: 61 YPQIQFDLVFNKEFELSHTSKTTSVFFSGYKVEQPIEGDEMDLDSEDEEEELNIPVIKEN 120
Query: 823 GKGDGKEEQKNQEKAVAATAXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXX 644
GK DGKEEQKNQEKAVAATA
Sbjct: 121 GKADGKEEQKNQEKAVAATASKSSLGLEKKSKDDSDDSDEDESDDSDEDDSDDSDEGEGL 180
Query: 643 XXXXXXXXXXXXXXXXXXXXXXXXXXXXXEAGKKRGAENALKTPLSDKKAKVATPPAQKT 464
EAGKKRGAENALKTPLSDKKAKVATPPAQKT
Sbjct: 181 SPDEGDDDSSDEDDTSDDDEEETPTPKKPEAGKKRGAENALKTPLSDKKAKVATPPAQKT 240
Query: 463 GGKKGATHVATPHPAKGKTPANNDKSTEKSPKSGGSVPCKSCSKTFNSEMALQAHSKAKH 284
GGKKGATHVATPHPAKGKTPANNDK TEKSPKSGGSVPCKSCSKTFNSEMALQAHSKAKH
Sbjct: 241 GGKKGATHVATPHPAKGKTPANNDKLTEKSPKSGGSVPCKSCSKTFNSEMALQAHSKAKH 300
Query: 283 GAK 275
GAK
Sbjct: 301 GAK 303
>sptr|O24591|O24591 Nucleolar histone deacetylase HD2-P39.
Length = 307
Score = 209 bits (532), Expect = 7e-53
Identities = 104/138 (75%), Positives = 118/138 (85%)
Frame = -1
Query: 1183 MEFWGLEVKPGSTVKCEPGHGFILHVSQAALGESKKSDSALMYVKVDDKKLAIGTLSIDK 1004
MEFWGLEVKPGSTVKCEPG+GF+LH+SQAALGESKKSD+ALMYVK+DD+KLAIGTLS+DK
Sbjct: 1 MEFWGLEVKPGSTVKCEPGYGFVLHLSQAALGESKKSDNALMYVKIDDQKLAIGTLSVDK 60
Query: 1003 YPQIQFDLVFNKEFELSHTSKTTSVFFSGYKVEQPIXXXXXXXXXXXXXXELNIPVIKEN 824
P IQFDL+F+KEFELSHTSKTTSVFF+GYKVEQP ELN+PV+KEN
Sbjct: 61 NPHIQFDLIFDKEFELSHTSKTTSVFFTGYKVEQPFEEDEMDLDSEDEDEELNVPVVKEN 120
Query: 823 GKGDGKEEQKNQEKAVAA 770
GK D K +QK+QEKAVAA
Sbjct: 121 GKADEK-KQKSQEKAVAA 137
Score = 135 bits (341), Expect = 1e-30
Identities = 73/96 (76%), Positives = 80/96 (83%), Gaps = 5/96 (5%)
Frame = -1
Query: 550 GKKRGAENA-LKTPLSDKKAKVATPPAQKTGGKKGAT-HVATPHPAKGKTPANNDKSTEK 377
GKKR AE++ LKTPLSDKKAKVATP +QKTGGKKGA HVATPHPAKGKT NNDKS +
Sbjct: 206 GKKRPAESSVLKTPLSDKKAKVATPSSQKTGGKKGAAVHVATPHPAKGKTIVNNDKSVKS 265
Query: 376 ---SPKSGGSVPCKSCSKTFNSEMALQAHSKAKHGA 278
+PKSGGSVPCK CSK+F SE ALQAHS+AK GA
Sbjct: 266 PKSAPKSGGSVPCKPCSKSFISETALQAHSRAKMGA 301
>sptr|Q9M4U4|Q9M4U4 Histone deacetylase 2 isoform c.
Length = 300
Score = 207 bits (528), Expect = 2e-52
Identities = 103/138 (74%), Positives = 117/138 (84%)
Frame = -1
Query: 1183 MEFWGLEVKPGSTVKCEPGHGFILHVSQAALGESKKSDSALMYVKVDDKKLAIGTLSIDK 1004
MEFWGLEVKPGSTVKCEPG+GF+LH+SQAALGESKKSD+ALMYVK+DD+KLAIGTLS+DK
Sbjct: 1 MEFWGLEVKPGSTVKCEPGYGFVLHLSQAALGESKKSDNALMYVKIDDQKLAIGTLSVDK 60
Query: 1003 YPQIQFDLVFNKEFELSHTSKTTSVFFSGYKVEQPIXXXXXXXXXXXXXXELNIPVIKEN 824
P IQFDL+F+KEFELSHTSKTTSVFF+GYKVEQP ELN+P +KEN
Sbjct: 61 NPHIQFDLIFDKEFELSHTSKTTSVFFTGYKVEQPFEEDEMDLDSEDEDEELNVPAVKEN 120
Query: 823 GKGDGKEEQKNQEKAVAA 770
GK D K +QK+QEKAVAA
Sbjct: 121 GKADEK-KQKSQEKAVAA 137
Score = 133 bits (335), Expect = 5e-30
Identities = 74/96 (77%), Positives = 78/96 (81%), Gaps = 4/96 (4%)
Frame = -1
Query: 550 GKKRGAENA-LKTPLSDKKAKVATPPAQKTGGKKG-ATHVATPHPAKGKTPANNDKSTEK 377
GKKR AE++ LKTPLSDKKAKVATP +QKTGGKKG A HVATPHPAKGKT NNDKS K
Sbjct: 206 GKKRAAESSVLKTPLSDKKAKVATPSSQKTGGKKGAAVHVATPHPAKGKTIVNNDKSV-K 264
Query: 376 SPKSG--GSVPCKSCSKTFNSEMALQAHSKAKHGAK 275
SPKS VPCKSCSK+F SE A QAHSKAKHG K
Sbjct: 265 SPKSAPKSGVPCKSCSKSFISETAPQAHSKAKHGGK 300
>sptr|Q9M4T5|Q9M4T5 Putative histone deacetylase HD2.
Length = 297
Score = 164 bits (414), Expect = 3e-39
Identities = 92/140 (65%), Positives = 100/140 (71%), Gaps = 6/140 (4%)
Frame = -1
Query: 1183 MEFWGLEVKPGSTVKCEPGHGFILHVSQAALGESKK-SDSALMYVKVDDKKLAIGTLSID 1007
MEFWGLEVKPG TVKCEP LH+SQAALGESKK SD+A+MYVK DD+KL IGTLS D
Sbjct: 1 MEFWGLEVKPGQTVKCEPEDERFLHLSQAALGESKKGSDNAVMYVKTDDQKLVIGTLSAD 60
Query: 1006 KYPQIQFDLVFNKEFELSHTSKTTSVFFSGYKVEQPIXXXXXXXXXXXXXXELN----IP 839
K+PQIQFDLVF+KEFELSHTSKT SVFFSGYKV QP E IP
Sbjct: 61 KFPQIQFDLVFDKEFELSHTSKTASVFFSGYKVSQPAEEDEMDFDSEEVEDEEEEEKIIP 120
Query: 838 VIKENGKGDGKE-EQKNQEK 782
+ NGK +GKE EQK Q K
Sbjct: 121 APRANGKVEGKENEQKKQGK 140
Score = 131 bits (330), Expect = 2e-29
Identities = 72/95 (75%), Positives = 77/95 (81%), Gaps = 3/95 (3%)
Frame = -1
Query: 550 GKKRGAENALKTPLSDKKAKVATPPAQKTGGKKGATHVATPHPAK--GKTPANNDKSTEK 377
GK++ AE LKTP SDKKAK+ATP QKTG KKG HVATPHPAK KTP N DKS EK
Sbjct: 205 GKRKVAEIVLKTPSSDKKAKIATPSGQKTGDKKGV-HVATPHPAKQASKTPVN-DKSKEK 262
Query: 376 SPKSGG-SVPCKSCSKTFNSEMALQAHSKAKHGAK 275
SPKSGG S+ CKSCSKTFNSEMALQ+HSKAKH AK
Sbjct: 263 SPKSGGGSISCKSCSKTFNSEMALQSHSKAKHPAK 297
>sptr|Q94F81|Q94F81 HD2 type histone deacetylase HDA106.
Length = 286
Score = 124 bits (312), Expect = 2e-27
Identities = 60/97 (61%), Positives = 77/97 (79%), Gaps = 1/97 (1%)
Frame = -1
Query: 1183 MEFWGLEVKPGSTVKCEPGHGFILHVSQAALGESKK-SDSALMYVKVDDKKLAIGTLSID 1007
ME G EVKPG+TV C+ G G ++H+SQAALGESKK S++A++ V +DDKKL +GTLS++
Sbjct: 1 MEVGGQEVKPGATVSCKVGDGLVIHLSQAALGESKKASENAILSVNIDDKKLVLGTLSVE 60
Query: 1006 KYPQIQFDLVFNKEFELSHTSKTTSVFFSGYKVEQPI 896
K+PQI DLVF+K+FEL H SKT SVFF GYK P+
Sbjct: 61 KHPQISCDLVFDKDFELPHNSKTRSVFFRGYKSPVPL 97
>sptrnew|AAQ24532|AAQ24532 Histone deacetylase.
Length = 269
Score = 112 bits (279), Expect = 2e-23
Identities = 54/97 (55%), Positives = 70/97 (72%), Gaps = 2/97 (2%)
Frame = -1
Query: 1183 MEFWGLEVKPGSTVKCEPGHGFILHVSQAALGESKK--SDSALMYVKVDDKKLAIGTLSI 1010
MEFWG EVK G + +PG G +LH+SQA+LGE KK S+S + V +D KKL +GTL+
Sbjct: 1 MEFWGAEVKSGEPLTVQPGDGMVLHLSQASLGELKKDKSESVCLSVNIDGKKLVLGTLNS 60
Query: 1009 DKYPQIQFDLVFNKEFELSHTSKTTSVFFSGYKVEQP 899
+K PQ QFDLVF+++FELSH K+ SV+F GYK P
Sbjct: 61 EKVPQQQFDLVFDRDFELSHNLKSGSVYFFGYKATNP 97
Score = 96.7 bits (239), Expect = 7e-19
Identities = 50/90 (55%), Positives = 62/90 (68%)
Frame = -1
Query: 547 KKRGAENALKTPLSDKKAKVATPPAQKTGGKKGATHVATPHPAKGKTPANNDKSTEKSPK 368
K+R A++A KTP++DKKAK+ TP QKT GKKG HVATPHP+K K+PK
Sbjct: 189 KRRKADSATKTPVTDKKAKLTTP--QKTDGKKGGGHVATPHPSK---------QASKTPK 237
Query: 367 SGGSVPCKSCSKTFNSEMALQAHSKAKHGA 278
S GS CK C+++F SE AL +HSKAKH A
Sbjct: 238 SAGSHHCKPCNRSFGSEGALDSHSKAKHSA 267
>sptr|Q8LQJ1|Q8LQJ1 Histone deacetylase HD2-like protein.
Length = 688
Score = 103 bits (257), Expect = 5e-21
Identities = 47/99 (47%), Positives = 68/99 (68%)
Frame = -1
Query: 1207 FVSAPPPSMEFWGLEVKPGSTVKCEPGHGFILHVSQAALGESKKSDSALMYVKVDDKKLA 1028
FV +M FWG+ V+PG TV C+P F H+SQ AL + +++ ++ +VD K++
Sbjct: 384 FVPPMETTMGFWGVAVRPGETVMCDPPGEFYYHISQIALEPGELNENVQVFGEVDGKRIL 443
Query: 1027 IGTLSIDKYPQIQFDLVFNKEFELSHTSKTTSVFFSGYK 911
+GTLS++ PQ+ DLVF KEFEL HTSKT ++FFSGY+
Sbjct: 444 LGTLSVEHRPQLSIDLVFEKEFELLHTSKTYNIFFSGYQ 482
Score = 38.1 bits (87), Expect = 0.28
Identities = 24/57 (42%), Positives = 31/57 (54%), Gaps = 5/57 (8%)
Frame = -1
Query: 466 TGGKKGATHVATPHPAK--GKTPANNDKSTEKS--PKSGGSVPCKSCSK-TFNSEMA 311
TG + G HVATP+PAK KTP NND + + S P K K T N++M+
Sbjct: 619 TGKRSGYVHVATPYPAKQAKKTPVNNDMAKQSSGYVHVATPYPAKQAKKRTANNDMS 675
Score = 37.7 bits (86), Expect = 0.37
Identities = 26/70 (37%), Positives = 35/70 (50%), Gaps = 2/70 (2%)
Frame = -1
Query: 535 AENALKTPLSDKKAKVATPPAQKTGGKKGATHVATPHPAK--GKTPANNDKSTEKSPKSG 362
A+ A KTP+++ AK ++ G HVATP+PAK K ANND S +
Sbjct: 634 AKQAKKTPVNNDMAKQSS----------GYVHVATPYPAKQAKKRTANNDMS-----EHS 678
Query: 361 GSVPCKSCSK 332
CK C+K
Sbjct: 679 AGYACKPCNK 688
>sptr|Q9FVE6|Q9FVE6 Putative histone deacetylase.
Length = 245
Score = 99.0 bits (245), Expect = 1e-19
Identities = 50/93 (53%), Positives = 65/93 (69%), Gaps = 2/93 (2%)
Frame = -1
Query: 1183 MEFWGLEVKPGSTVKCEPGHGFILHVSQAALGE--SKKSDSALMYVKVDDKKLAIGTLSI 1010
MEFWG+EVK G V P G ++HVSQA+LGE +KK + ++VKV ++ L +GTLS
Sbjct: 1 MEFWGIEVKSGKPVTVTPEEGILIHVSQASLGECKNKKGEFVPLHVKVGNQNLVLGTLST 60
Query: 1009 DKYPQIQFDLVFNKEFELSHTSKTTSVFFSGYK 911
+ PQ+ DLVF+KEFELSHT SV+F GYK
Sbjct: 61 ENIPQLFCDLVFDKEFELSHTWGKGSVYFVGYK 93
Score = 67.8 bits (164), Expect = 3e-10
Identities = 44/92 (47%), Positives = 50/92 (54%)
Frame = -1
Query: 553 AGKKRGAENALKTPLSDKKAKVATPPAQKTGGKKGATHVATPHPAKGKTPANNDKSTEKS 374
+ KKR E K P+S KKAKVA P QKT KK KG AN +S
Sbjct: 170 SSKKRANETTPKAPVSAKKAKVAVTP-QKTDEKK-----------KGGKAAN------QS 211
Query: 373 PKSGGSVPCKSCSKTFNSEMALQAHSKAKHGA 278
PKS V C SC KTFNS AL++H+KAKH A
Sbjct: 212 PKSASQVSCGSCKKTFNSGNALESHNKAKHAA 243
>sptr|O22238|O22238 Putative histone deacetylase.
Length = 257
Score = 99.0 bits (245), Expect = 1e-19
Identities = 50/93 (53%), Positives = 65/93 (69%), Gaps = 2/93 (2%)
Frame = -1
Query: 1183 MEFWGLEVKPGSTVKCEPGHGFILHVSQAALGE--SKKSDSALMYVKVDDKKLAIGTLSI 1010
MEFWG+EVK G V P G ++HVSQA+LGE +KK + ++VKV ++ L +GTLS
Sbjct: 1 MEFWGIEVKSGKPVTVTPEEGILIHVSQASLGECKNKKGEFVPLHVKVGNQNLVLGTLST 60
Query: 1009 DKYPQIQFDLVFNKEFELSHTSKTTSVFFSGYK 911
+ PQ+ DLVF+KEFELSHT SV+F GYK
Sbjct: 61 ENIPQLFCDLVFDKEFELSHTWGKGSVYFVGYK 93
Score = 67.8 bits (164), Expect = 3e-10
Identities = 44/92 (47%), Positives = 50/92 (54%)
Frame = -1
Query: 553 AGKKRGAENALKTPLSDKKAKVATPPAQKTGGKKGATHVATPHPAKGKTPANNDKSTEKS 374
+ KKR E K P+S KKAKVA P QKT KK KG AN +S
Sbjct: 170 SSKKRANETTPKAPVSAKKAKVAVTP-QKTDEKK-----------KGGKAAN------QS 211
Query: 373 PKSGGSVPCKSCSKTFNSEMALQAHSKAKHGA 278
PKS V C SC KTFNS AL++H+KAKH A
Sbjct: 212 PKSASQVSCGSCKKTFNSGNALESHNKAKHAA 243
>sptr|Q9M4T4|Q9M4T4 Putative histone deacetylase HD2c
(AT5g03740/F17C15_160) (HDA11).
Length = 294
Score = 98.2 bits (243), Expect = 2e-19
Identities = 49/95 (51%), Positives = 65/95 (68%), Gaps = 2/95 (2%)
Frame = -1
Query: 1183 MEFWGLEVKPGSTVKCEPGHGFILHVSQAALGESKKS--DSALMYVKVDDKKLAIGTLSI 1010
MEFWG+EVK G + +PG ++H+SQ ALGESK + + +YV V KL IGTLS
Sbjct: 1 MEFWGVEVKNGKPLHLDPGLDRLVHISQVALGESKNNVTEPIQLYVTVGSDKLLIGTLSH 60
Query: 1009 DKYPQIQFDLVFNKEFELSHTSKTTSVFFSGYKVE 905
+K+PQ+ ++V + F LSHT K SVFFSGYKV+
Sbjct: 61 EKFPQLSTEIVLERNFALSHTWKNGSVFFSGYKVD 95
Score = 91.3 bits (225), Expect = 3e-17
Identities = 51/97 (52%), Positives = 63/97 (64%), Gaps = 7/97 (7%)
Frame = -1
Query: 547 KKRGAE-NALKTPLSDKKAKVATPPAQKTGGKKGATHVATPHPAKGKTPANNDKST---- 383
KKR AE N+ K P S+KKAK TP QKT KK HVATPHP+K + ST
Sbjct: 198 KKRSAEPNSSKNPASNKKAKFVTP--QKTDSKKPHVHVATPHPSKQAGKNSGGGSTGETS 255
Query: 382 --EKSPKSGGSVPCKSCSKTFNSEMALQAHSKAKHGA 278
+++PKS G+ CKSC++TF SEM LQ+H+KAKH A
Sbjct: 256 KQQQTPKSAGAFGCKSCTRTFTSEMGLQSHTKAKHSA 292
>sptr|Q9LZR5|Q9LZR5 Histone deacetylase-like protein.
Length = 296
Score = 97.4 bits (241), Expect = 4e-19
Identities = 50/98 (51%), Positives = 65/98 (66%), Gaps = 2/98 (2%)
Frame = -1
Query: 1183 MEFWGLEVKPGSTVKCEPGHGFILHVSQAALGESKKS--DSALMYVKVDDKKLAIGTLSI 1010
MEFWG+EVK G + +PG ++H+SQ ALGESK + + +YV V KL IGTLS
Sbjct: 1 MEFWGVEVKNGKPLHLDPGLDRLVHISQVALGESKNNVTEPIQLYVTVGSDKLLIGTLSH 60
Query: 1009 DKYPQIQFDLVFNKEFELSHTSKTTSVFFSGYKVEQPI 896
+K+PQ+ ++V + F LSHT K SVFFSGYK E I
Sbjct: 61 EKFPQLSTEIVLERNFALSHTWKNGSVFFSGYKPEDLI 98
Score = 91.3 bits (225), Expect = 3e-17
Identities = 51/97 (52%), Positives = 63/97 (64%), Gaps = 7/97 (7%)
Frame = -1
Query: 547 KKRGAE-NALKTPLSDKKAKVATPPAQKTGGKKGATHVATPHPAKGKTPANNDKST---- 383
KKR AE N+ K P S+KKAK TP QKT KK HVATPHP+K + ST
Sbjct: 200 KKRSAEPNSSKNPASNKKAKFVTP--QKTDSKKPHVHVATPHPSKQAGKNSGGGSTGETS 257
Query: 382 --EKSPKSGGSVPCKSCSKTFNSEMALQAHSKAKHGA 278
+++PKS G+ CKSC++TF SEM LQ+H+KAKH A
Sbjct: 258 KQQQTPKSAGAFGCKSCTRTFTSEMGLQSHTKAKHSA 294
>sptr|Q8LJS2|Q8LJS2 Nucleolar histone deacetylase HD2-P39.
Length = 295
Score = 96.3 bits (238), Expect = 9e-19
Identities = 49/97 (50%), Positives = 67/97 (69%), Gaps = 5/97 (5%)
Frame = -1
Query: 1183 MEFWGLEVKPGSTVKCEPGHGF--ILHVSQAALGESKK---SDSALMYVKVDDKKLAIGT 1019
MEFWG+EVK G TV +P +H+SQ ALGE+KK ++ ++Y+KV ++K+ +GT
Sbjct: 1 MEFWGVEVKVGQTVTVDPMDPVDSYIHISQVALGEAKKDKPNEPVVLYLKVGEQKIVLGT 60
Query: 1018 LSIDKYPQIQFDLVFNKEFELSHTSKTTSVFFSGYKV 908
LS D P + DLV + + ELSHTSK+ SVFF GYKV
Sbjct: 61 LSRDGIPHLSLDLVLDSDSELSHTSKSASVFFCGYKV 97
Score = 94.0 bits (232), Expect = 4e-18
Identities = 53/94 (56%), Positives = 65/94 (69%), Gaps = 2/94 (2%)
Frame = -1
Query: 550 GKKRGAENALKTPLSDKKAKVATPPAQKTGGKKGATHVATPHPAK--GKTPANNDKSTEK 377
GKKR E+A KTP+S KKAK ATP +KT GKK + HVATPHP+K GKTP N+ K +
Sbjct: 208 GKKRPNESAAKTPISAKKAKTATP--EKTDGKK-SVHVATPHPSKKGGKTP-NSTKG--Q 261
Query: 376 SPKSGGSVPCKSCSKTFNSEMALQAHSKAKHGAK 275
+P S G + C SC K+F +E LQ H KAKHG +
Sbjct: 262 TPNSAGQLSCASCKKSFTNEAGLQQHKKAKHGGQ 295
>sptr|O49209|O49209 Putative histone deacetylase.
Length = 305
Score = 92.8 bits (229), Expect = 1e-17
Identities = 48/92 (52%), Positives = 62/92 (67%), Gaps = 1/92 (1%)
Frame = -1
Query: 1183 MEFWGLEVKPGSTVKCEPGHGFILHVSQAALGESKKS-DSALMYVKVDDKKLAIGTLSID 1007
MEFWG+ V P + K P ++H+SQA+L + KS +S ++ V V KL IGTLS D
Sbjct: 1 MEFWGVAVTPKNATKVTPEEDSLVHISQASLDCTVKSGESVVLSVTVGGAKLVIGTLSQD 60
Query: 1006 KYPQIQFDLVFNKEFELSHTSKTTSVFFSGYK 911
K+PQI FDLVF+KEFELSH+ +V F GYK
Sbjct: 61 KFPQISFDLVFDKEFELSHSGTKANVHFIGYK 92
Score = 62.0 bits (149), Expect = 2e-08
Identities = 44/82 (53%), Positives = 49/82 (59%), Gaps = 6/82 (7%)
Frame = -1
Query: 547 KKRGAENALKTPLSDKKAKVATPPA---QKTGGKKGATHVATPHPAK--GKTPANNDKST 383
KKR E+ KTP+S KKAK A PA QKT KKG H ATPHPAK GK+P N
Sbjct: 208 KKRPNESVSKTPVSGKKAKPAAAPASTPQKTEKKKGG-HTATPHPAKKGGKSPVN----A 262
Query: 382 EKSPKSGG-SVPCKSCSKTFNS 320
+SPKSGG S + K FNS
Sbjct: 263 NQSPKSGGQSSGGNNNKKPFNS 284
>sptr|Q9FNJ6|Q9FNJ6 Histone deacetylase-like protein
(AT5g22650/MDJ22_7).
Length = 306
Score = 92.8 bits (229), Expect = 1e-17
Identities = 48/92 (52%), Positives = 62/92 (67%), Gaps = 1/92 (1%)
Frame = -1
Query: 1183 MEFWGLEVKPGSTVKCEPGHGFILHVSQAALGESKKS-DSALMYVKVDDKKLAIGTLSID 1007
MEFWG+ V P + K P ++H+SQA+L + KS +S ++ V V KL IGTLS D
Sbjct: 1 MEFWGVAVTPKNATKVTPEEDSLVHISQASLDCTVKSGESVVLSVTVGGAKLVIGTLSQD 60
Query: 1006 KYPQIQFDLVFNKEFELSHTSKTTSVFFSGYK 911
K+PQI FDLVF+KEFELSH+ +V F GYK
Sbjct: 61 KFPQISFDLVFDKEFELSHSGTKANVHFIGYK 92
Score = 62.8 bits (151), Expect = 1e-08
Identities = 43/82 (52%), Positives = 48/82 (58%), Gaps = 6/82 (7%)
Frame = -1
Query: 547 KKRGAENALKTPLSDKKAKVATPPA---QKTGGKKGATHVATPHPAK--GKTPANNDKST 383
KKR E+ KTP+S KKAK A PA QKT KK H ATPHPAK GK+P N
Sbjct: 208 KKRPNESVSKTPVSGKKAKPAAAPASTPQKTEEKKKGGHTATPHPAKKGGKSPVN----A 263
Query: 382 EKSPKSGG-SVPCKSCSKTFNS 320
+SPKSGG S + K FNS
Sbjct: 264 NQSPKSGGQSSGGNNNKKPFNS 285
>sptrnew|AAP40362|AAP40362 Hypothetical protein At2g27840.
Length = 203
Score = 78.2 bits (191), Expect = 2e-13
Identities = 43/96 (44%), Positives = 58/96 (60%), Gaps = 5/96 (5%)
Frame = -1
Query: 1183 MEFWGLEVKPGSTVKCEPGHGFILHVSQAALGE---SKKSDSALMYVKV--DDKKLAIGT 1019
MEFWG+E+KPG K GF++H SQ LG+ KK ++ +YVK+ D+ IG
Sbjct: 1 MEFWGIEIKPGKPFKVIQKDGFMVHASQVTLGDVEKVKKDETFAVYVKIGDDENGFMIGN 60
Query: 1018 LSIDKYPQIQFDLVFNKEFELSHTSKTTSVFFSGYK 911
LS K+PQ DL EFE+SH S T+SV+ GY+
Sbjct: 61 LS-QKFPQFSIDLYLGHEFEISHNS-TSSVYLIGYR 94
>sptr|Q9M4T3|Q9M4T3 Putative histone deacetylase HD2d (Hypothetical
protein At2g27840).
Length = 203
Score = 78.2 bits (191), Expect = 2e-13
Identities = 43/96 (44%), Positives = 58/96 (60%), Gaps = 5/96 (5%)
Frame = -1
Query: 1183 MEFWGLEVKPGSTVKCEPGHGFILHVSQAALGE---SKKSDSALMYVKV--DDKKLAIGT 1019
MEFWG+E+KPG K GF++H SQ LG+ KK ++ +YVK+ D+ IG
Sbjct: 1 MEFWGIEIKPGKPFKVIQKDGFMVHASQVTLGDVEKVKKDETFAVYVKIGDDENGFMIGN 60
Query: 1018 LSIDKYPQIQFDLVFNKEFELSHTSKTTSVFFSGYK 911
LS K+PQ DL EFE+SH S T+SV+ GY+
Sbjct: 61 LS-QKFPQFSIDLYLGHEFEISHNS-TSSVYLIGYR 94
>sptr|Q8LQJ0|Q8LQJ0 P0456E05.12 protein.
Length = 1627
Score = 54.7 bits (130), Expect = 3e-06
Identities = 32/83 (38%), Positives = 50/83 (60%), Gaps = 5/83 (6%)
Frame = -1
Query: 508 SDKKAKVATPPAQKTGGKKGATHVA---TPHPAK--GKTPANNDKSTEKSPKSGGSVPCK 344
SD++ ++ P G+T + HPAK GKTPA + K +++P+S G+ CK
Sbjct: 1546 SDEEDEIPEKPESSKLTAAGSTLSSGDKNTHPAKEDGKTPAIS-KPNKETPESSGTHACK 1604
Query: 343 SCSKTFNSEMALQAHSKAKHGAK 275
CSK F+S+ +L++H KA+H AK
Sbjct: 1605 YCSKAFSSDKSLRSHQKARHPAK 1627
>sptr|Q9ZUY5|Q9ZUY5 Hypothetical 20.1 kDa protein.
Length = 181
Score = 52.0 bits (123), Expect = 2e-05
Identities = 31/74 (41%), Positives = 44/74 (59%), Gaps = 5/74 (6%)
Frame = -1
Query: 1117 ILHVSQAALGES---KKSDSALMYVKV--DDKKLAIGTLSIDKYPQIQFDLVFNKEFELS 953
++H SQ LG+ KK ++ +YVK+ D+ IG LS K+PQ DL EFE+S
Sbjct: 1 MVHASQVTLGDVEKVKKDETFAVYVKIGDDENGFMIGNLS-QKFPQFSIDLYLGHEFEIS 59
Query: 952 HTSKTTSVFFSGYK 911
H S T+SV+ GY+
Sbjct: 60 HNS-TSSVYLIGYR 72
>sw|Q08864|H11_VOLCA Histone H1-I.
Length = 260
Score = 41.6 bits (96), Expect = 0.025
Identities = 33/88 (37%), Positives = 45/88 (51%), Gaps = 1/88 (1%)
Frame = -1
Query: 547 KKRGAENALKTPLSDKKAKVATPPAQKTGGKKGATHVATPHP-AKGKTPANNDKSTEKSP 371
KK E TP ++K K ATP K+ GKK ATP P A K+PA D +K+
Sbjct: 180 KKEKVEKKKATPKAEKPKKAATP---KSAGKK----KATPKPKAAPKSPAKKDAKPKKAT 232
Query: 370 KSGGSVPCKSCSKTFNSEMALQAHSKAK 287
S + P K+ +K ++ A +A SK K
Sbjct: 233 PSKKAAPKKAPAKK-STPKAKEAKSKGK 259
>sptr|Q7ZZ75|Q7ZZ75 SI:dZ44O19.5 (Novel zinc finger protein (5
domains)).
Length = 595
Score = 41.2 bits (95), Expect = 0.033
Identities = 34/119 (28%), Positives = 52/119 (43%), Gaps = 7/119 (5%)
Frame = -1
Query: 529 NALKTPLSDKKAKVATPPA------QKTGGKKGATHV-ATPHPAKGKTPANNDKSTEKSP 371
+AL +DKK + PPA K G KK + A PH +ND+ +
Sbjct: 164 SALHPSNTDKKEDGSIPPAVVMPIPSKRGRKKKSMMPRAGPHNTNDPYDLSNDEDEHQGK 223
Query: 370 KSGGSVPCKSCSKTFNSEMALQAHSKAKHGAK*VAGPTESMKEKHGGQVSRRSIRVHEG 194
S C+ C+ TF S+ +Q H+K+ AK P S + +++ IR+H G
Sbjct: 224 DGNKSYRCRMCAVTFFSKSDMQIHAKSHTEAKPHKCPHCSKSFANSSYLAQH-IRIHSG 281
>sptrnew|AAH56460|AAH56460 Hypothetical protein (Fragment).
Length = 879
Score = 39.7 bits (91), Expect = 0.096
Identities = 33/127 (25%), Positives = 52/127 (40%), Gaps = 11/127 (8%)
Frame = -1
Query: 553 AGKKRGAENALKTPLSDKKAKVATPPAQKTGG-----------KKGATHVATPHPAKGKT 407
+G + + L TP++ K + TP A G ++G T + KT
Sbjct: 451 SGSEEVRKKVLATPVTVSK-EAPTPVAHPAPGGPEEQWQRAIHERGEAVCPTCNVVTRKT 509
Query: 406 PANNDKSTEKSPKSGGSVPCKSCSKTFNSEMALQAHSKAKHGAK*VAGPTESMKEKHGGQ 227
K E K ++ C+ C K F S+ L H+ A+H AK PT++ + G Q
Sbjct: 510 LVGLKKHMEVCHKLQEALKCQHCRKQFKSKAGLNYHTMAEHSAK----PTDAEASEGGEQ 565
Query: 226 VSRRSIR 206
R +R
Sbjct: 566 EERERLR 572
>sptrnew|AAH53361|AAH53361 Hypothetical protein.
Length = 460
Score = 39.7 bits (91), Expect = 0.096
Identities = 34/130 (26%), Positives = 52/130 (40%), Gaps = 15/130 (11%)
Frame = -1
Query: 538 GAENALKTPLSDKKAKVATPPAQKTGGKKGATHVATPHPAKGKT---------PANND-- 392
G A + D KVA+ ++ GG G P P +G+ P ND
Sbjct: 97 GLMTAALQVVPDLSKKVASTLTEEGGGGGGGGGSVAPKPPRGRKKKRMLESGLPEMNDPY 156
Query: 391 ----KSTEKSPKSGGSVPCKSCSKTFNSEMALQAHSKAKHGAK*VAGPTESMKEKHGGQV 224
+ + K G + C+ CS TF S+ +Q HSK+ K P S + +
Sbjct: 157 VLSPEDDDDHQKDGKTYRCRMCSLTFYSKSEMQIHSKSHTETKPHKCPHCSKTFANSSYL 216
Query: 223 SRRSIRVHEG 194
++ IR+H G
Sbjct: 217 AQH-IRIHSG 225
>sptr|Q93ZG9|Q93ZG9 AT4g25340/T30C3_20.
Length = 477
Score = 39.7 bits (91), Expect = 0.096
Identities = 29/94 (30%), Positives = 46/94 (48%), Gaps = 4/94 (4%)
Frame = -1
Query: 1183 MEFWGLEVKPGSTVKCEP-GHGFILHVSQAALGESKKSDSALMYVKVDDK-KLAIGTLSI 1010
M FWGLEVKPG P +HV+QA LG + +++ + DK +A+ +L
Sbjct: 1 MGFWGLEVKPGKPQAYNPKNEQGKIHVTQATLGTGLSKEKSVIQCSIGDKAPIALCSLLP 60
Query: 1009 DKYPQIQFDLVFNKEFELSHTSKT--TSVFFSGY 914
+K +L F+ + E + T S+ SG+
Sbjct: 61 NKIECCPLNLEFDDDDEPVEFTVTGDRSIHLSGF 94
>sw|P56270|MAZ_HUMAN Myc-associated zinc finger protein (MAZI)
(Purine-binding transcription factor) (Pur-1) (ZF87)
(ZIF87).
Length = 477
Score = 39.7 bits (91), Expect = 0.096
Identities = 28/90 (31%), Positives = 38/90 (42%)
Frame = -1
Query: 514 PLSDKKAKVATPPAQKTGGKKGATHVATPHPAKGKTPANNDKSTEKSPKSGGSVPCKSCS 335
P+S A+ A P + T AT V P P + + EK KS G C C+
Sbjct: 139 PVSAPAAEAAPPASAATIAAAAATAVVAPTSTVAVAPVAS--ALEKKTKSKGPYICALCA 196
Query: 334 KTFNSEMALQAHSKAKHGAK*VAGPTESMK 245
K F + L+ H GAK P+ +MK
Sbjct: 197 KEFKNGYNLRRHEAIHTGAKAGRVPSGAMK 226
>sptr|Q8NFN7|Q8NFN7 Serum amyloid A activating factor 2.
Length = 493
Score = 39.7 bits (91), Expect = 0.096
Identities = 28/90 (31%), Positives = 38/90 (42%)
Frame = -1
Query: 514 PLSDKKAKVATPPAQKTGGKKGATHVATPHPAKGKTPANNDKSTEKSPKSGGSVPCKSCS 335
P+S A+ A P + T AT V P P + + EK KS G C C+
Sbjct: 139 PVSAPAAEAAPPASAATIAAAAATAVVAPTSTVAVAPVAS--ALEKKTKSKGPYICALCA 196
Query: 334 KTFNSEMALQAHSKAKHGAK*VAGPTESMK 245
K F + L+ H GAK P+ +MK
Sbjct: 197 KEFKNGYNLRRHEAIHTGAKAGRVPSGAMK 226
>sptrnew|EAA32141|EAA32141 Hypothetical protein ( (AL451013)
probable nucleotide exsicion repair protein RAD16.
Length = 1079
Score = 39.7 bits (91), Expect = 0.096
Identities = 26/86 (30%), Positives = 36/86 (41%), Gaps = 4/86 (4%)
Frame = -1
Query: 547 KKRGAENALKTPLSDKKAKVATPPAQKTGGKKGAT----HVATPHPAKGKTPANNDKSTE 380
K RG TP A PA T G + AT H ATP P+ K + +ST+
Sbjct: 25 KPRGRPRKSATPAVSATASSVPTPATYTSGSEYATPLTSHAATPTPSLLKENVRSTRSTQ 84
Query: 379 KSPKSGGSVPCKSCSKTFNSEMALQA 302
+PK ++P S T + + A
Sbjct: 85 SAPKIEVAIPVLKASHTMQTTLRSSA 110
>sptr|Q9HE48|Q9HE48 Probable nucleotide exsicion repair protein
RAD16.
Length = 1079
Score = 39.7 bits (91), Expect = 0.096
Identities = 26/86 (30%), Positives = 36/86 (41%), Gaps = 4/86 (4%)
Frame = -1
Query: 547 KKRGAENALKTPLSDKKAKVATPPAQKTGGKKGAT----HVATPHPAKGKTPANNDKSTE 380
K RG TP A PA T G + AT H ATP P+ K + +ST+
Sbjct: 25 KPRGRPRKSATPAVSATASSVPTPATYTSGSEYATPLTSHAATPTPSLLKENVRSTRSTQ 84
Query: 379 KSPKSGGSVPCKSCSKTFNSEMALQA 302
+PK ++P S T + + A
Sbjct: 85 SAPKIEVAIPVLKASHTMQTTLRSSA 110
>sw|Q8TF68|Z384_HUMAN Zinc finger protein 384 (Nuclear matrix
transcription factor 4) (CAG repeat protein 1).
Length = 576
Score = 39.3 bits (90), Expect = 0.13
Identities = 32/119 (26%), Positives = 49/119 (41%), Gaps = 15/119 (12%)
Frame = -1
Query: 505 DKKAKVATPPAQKTGGKKGATHVATPHPAKGKT---------PANND------KSTEKSP 371
D KVA+ ++ GG G P P +G+ P ND + +
Sbjct: 163 DLSKKVASTLTEEGGGGGGGGGSVAPKPPRGRKKKRMLESGLPEMNDPYVLSPEDDDDHQ 222
Query: 370 KSGGSVPCKSCSKTFNSEMALQAHSKAKHGAK*VAGPTESMKEKHGGQVSRRSIRVHEG 194
K G + C+ CS TF S+ +Q HSK+ K P S + +++ IR+H G
Sbjct: 223 KDGKTYRCRMCSLTFYSKSEMQIHSKSHTETKPHKCPHCSKTFANSSYLAQH-IRIHSG 280
>sptr|Q9VJN5|Q9VJN5 CG4148 protein (LD22579P).
Length = 470
Score = 39.3 bits (90), Expect = 0.13
Identities = 28/92 (30%), Positives = 42/92 (45%), Gaps = 17/92 (18%)
Frame = -1
Query: 505 DKKAKVATPPAQKTGGK-KGA---------THVATPHPAKGKTPANNDKSTEKSPKSGGS 356
D+ V+ PP ++ G+ KG+ +V + P +DK++E S GS
Sbjct: 204 DEDEAVSPPPLKRKRGRPKGSGKQKNVDDSDNVTSREPDDNAKSKQDDKTSELSMSPHGS 263
Query: 355 -------VPCKSCSKTFNSEMALQAHSKAKHG 281
PCK C++TF S MAL+ H HG
Sbjct: 264 QSSNFVDYPCKICNETFMSFMALRRHKHDMHG 295
>sptr|Q9N6X3|Q9N6X3 L(2)35Ea protein.
Length = 418
Score = 39.3 bits (90), Expect = 0.13
Identities = 28/92 (30%), Positives = 42/92 (45%), Gaps = 17/92 (18%)
Frame = -1
Query: 505 DKKAKVATPPAQKTGGK-KGA---------THVATPHPAKGKTPANNDKSTEKSPKSGGS 356
D+ V+ PP ++ G+ KG+ +V + P +DK++E S GS
Sbjct: 152 DEDEAVSPPPLKRKRGRPKGSGKQKNVDDSDNVTSREPDDNAKSKQDDKTSELSMSPHGS 211
Query: 355 -------VPCKSCSKTFNSEMALQAHSKAKHG 281
PCK C++TF S MAL+ H HG
Sbjct: 212 QSSNFVDYPCKICNETFMSFMALRRHKHDMHG 243
Database: /db/trembl-ebi/tmp/swall
Posted date: Sep 26, 2003 8:29 PM
Number of letters in database: 406,886,216
Number of sequences in database: 1,272,877
Lambda K H
0.318 0.135 0.401
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 914,259,640
Number of Sequences: 1272877
Number of extensions: 20806147
Number of successful extensions: 75674
Number of sequences better than 10.0: 30
Number of HSP's better than 10.0 without gapping: 64880
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 75301
length of database: 406,886,216
effective HSP length: 126
effective length of database: 246,503,714
effective search space used: 75183632770
frameshift window, decay const: 50, 0.1
T: 12
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)

